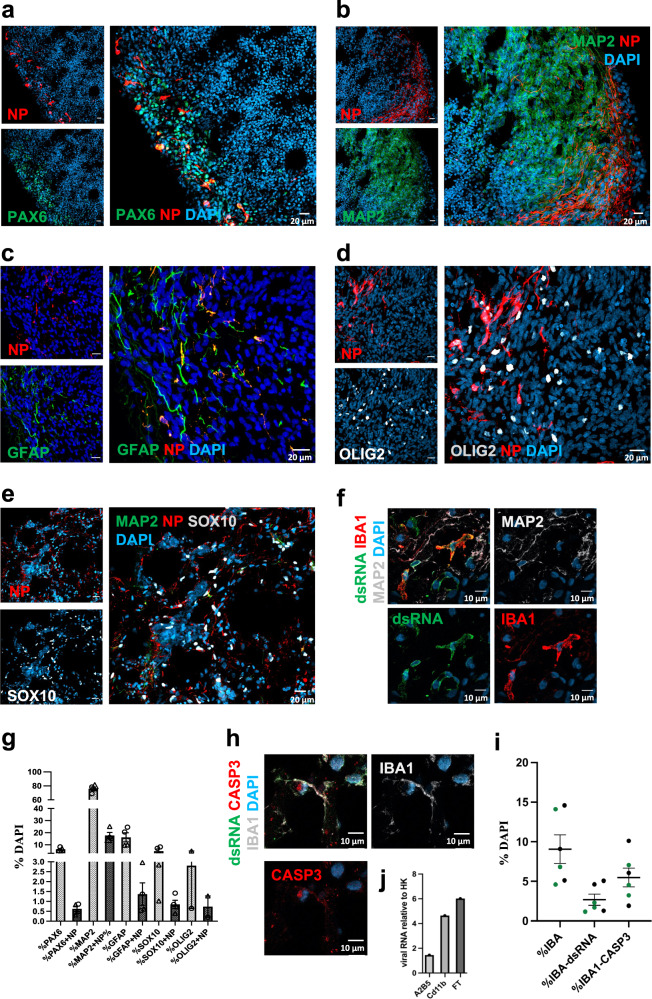
Fig. 3. Cell type specific SARS-CoV-2 infection in brain organoids with innately developing microglia.
Confocal images (40×) at 72hpi (MOI 0.3) showing colocalization of viral nucleoprotein (NP) with a neuronal precursors (PAX6), b neurons (MAP2), and a heterogenous population of progenitors marked by c GFAP d OLIG2 and e schwann cell precursors (SOX10). Scale bars for representative images a–e: 20 μM. Confocal images (40×) showing colocalization of organoid grown microglia (IBA1) with f dsRNA (clone 9D5, for viral presence). Nuclei stained with DAPI. Scale bar for f: 10 μM. g Distribution of NP+ cells amongst different cell-type markers, expressed as percentage of DAPI from two infected organoids at DIV 56 (corresponding data points indicated by circles and triangles). Each data point is an average of four FOVs representing different areas on the same section. Error bars indicate SEM. h Confocal images (40×) showing colocalization of organoid-grown microglia (IBA1) with CASP3 (obtained from one donor). Nuclei stained with DAPI. Scale bar for h: 10 μM. i Quantification of dsRNA and CASP3 presence in IBA1+ cells expressed as percentage of DAPI from two infected organoids at DIV 56 j qPCR on cell lysate fractions obtained after magnetic cell sorting (MACS) with CD11b beads, (also shown A2B5 radial glia enriched fraction) and flowthrough (FT), on infected organoids (n = 2, pooled) showing presence of viral N gene copies normalized to two housekeeping host genes. Center values represent means and error bars represent S.E.M.