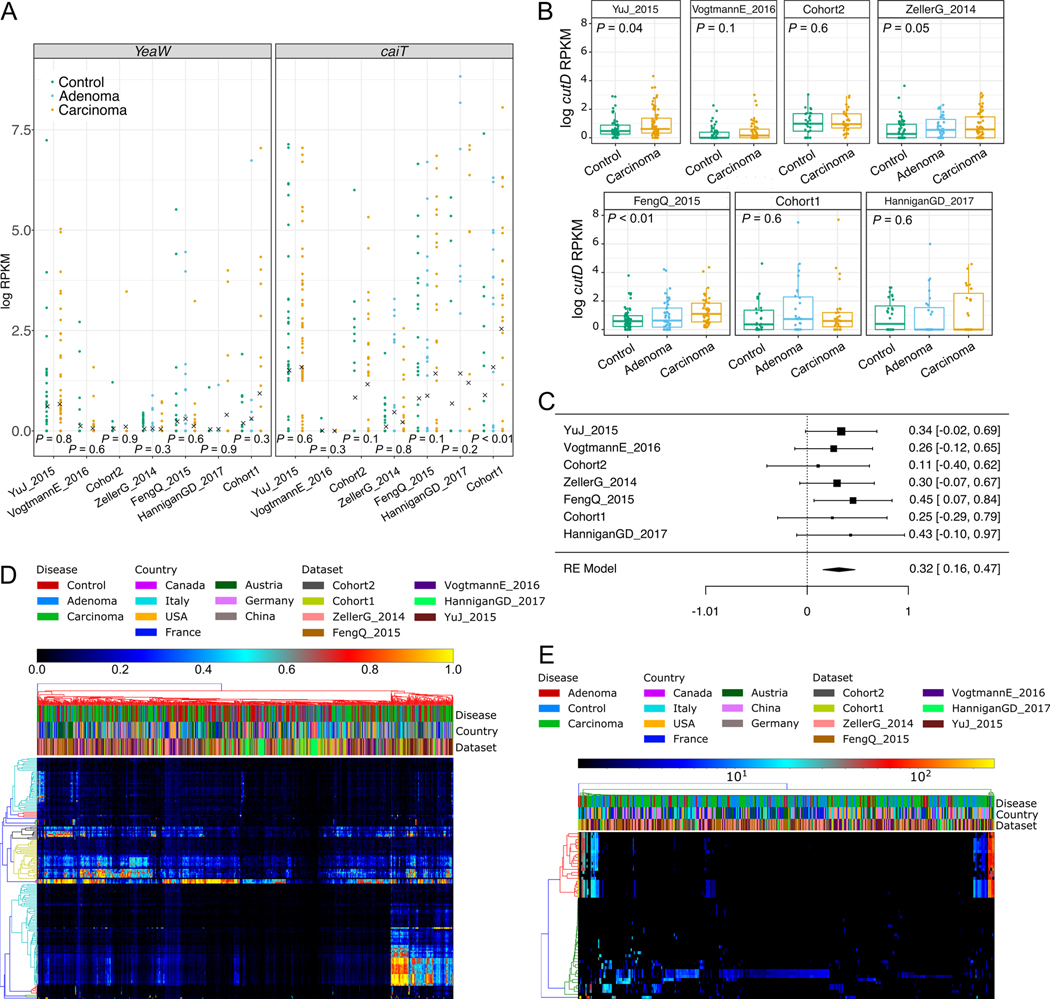
Fig. 9. Metagenomic analysis of genes involved in the TMA-synthesis pathway.
(A) ShortBRED analysis of yeaW and caiT gene abundances. Points represent the log of reads per kilobase per million mapped reads (RPKM) for each sample and crosses represent mean values per group/dataset. (B) ShortBRED analysis of cutD gene abundances. Boxplots reports the RKPM abundances obtained using ShortBRED for the gene of the activating TMA-lyase enzyme cutD. P-values were calculated by two-tailed Wilcoxon rank-sum tests comparing values between controls and carcinomas for each dataset. (C) Forest plot showing effect sizes calculated using a meta-analysis of standardized mean differences and a random effects model on cutD RPKM abundances between carcinomas and controls. (D) Breadth of coverage of cutC gene sequence clusters across CRC datasets. (E) Depth of coverage of cutC gene sequence clusters across CRC datasets..