Figure 3. Mammary tumor-induced de-differentiated adipocytes contribute to tumor microenvironment concerning inflammation and ECM remodeling.
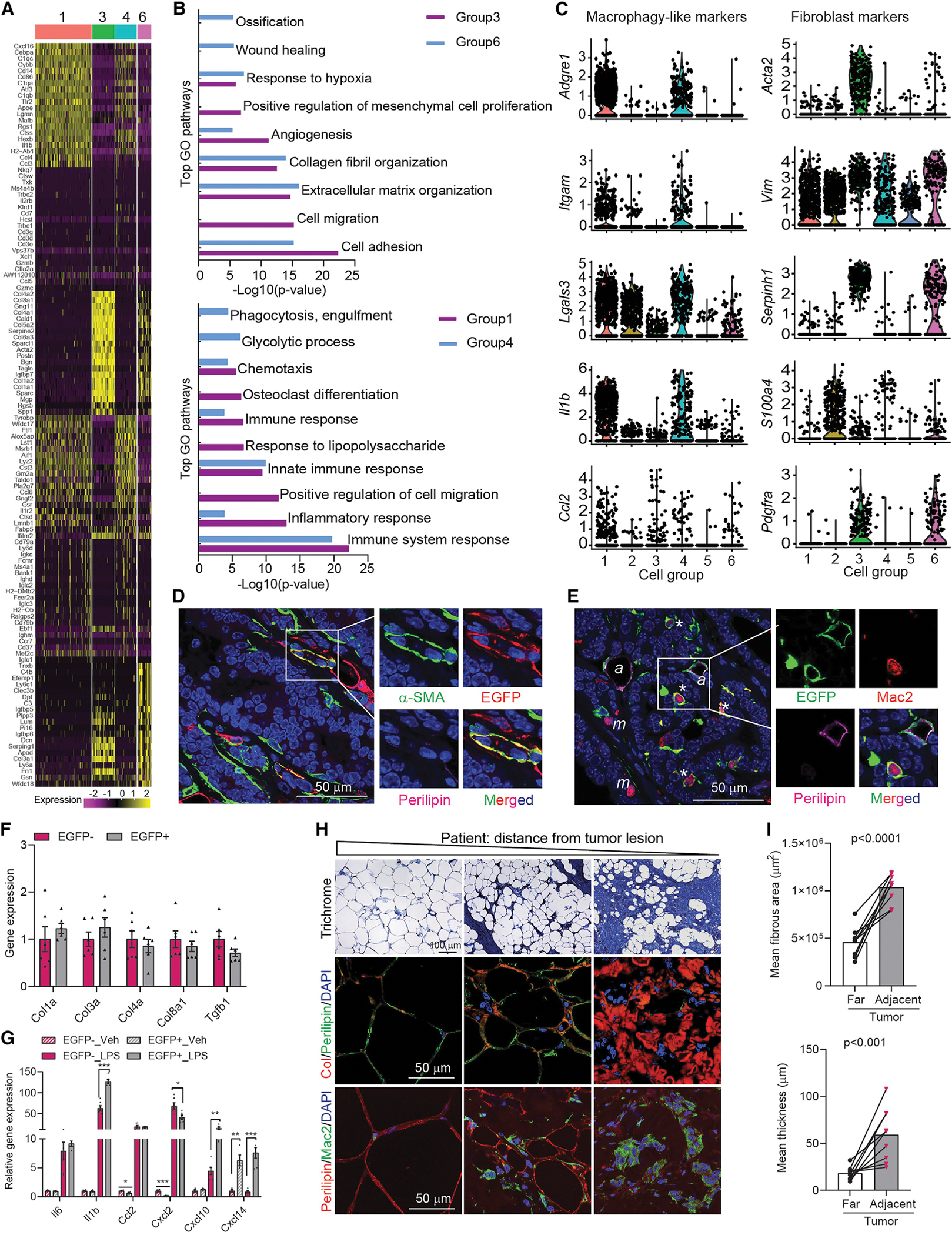
(A) Heat map of top 20 genes across different cell groups from allograft (Allo)-EGFP + cells.
(B) Gene ontology (GO) pathway enrichment analysis.
(C) Representative macrophage and fibroblast markers genes. The y-axis is the log-scale normalized read count.
(D) Co-staining of α-SMA (green), EGFP (red), and Perilipin (magenta) of PyMTChaser-tumor. Scale bar, 50 μm.
(E) Co-staining of EGFP (green), Mac2 (red), and Perilipin (magenta) of PyMTChaser-tumor. ‘*’ indicates macrophage-like cells (EGFP+/Perilipin−/Mac2+); ‘a’ indicates adipocytes (EGFP+/Perilipin+/Mac2−); ‘m’ indicates macrophages (EGFP−/Perilipin−/Mac2+). Scale bar, 50 μm.
(F) mRNA levels of genes related to ECM remodeling in cultures of de-differentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) isolated from AlloChaser-tumor (n = 6).
(G) mRNA levels of cytokines/chemokines in cultures of EGFP+ and EGFP− cells treated with vehicle (PBS) or LPS (100 ng/mL) for 3 h (n = 6).
(H) Trichrome, collagen (Col), and Mac2 staining indicates enhanced fibrosis, collagen production and inflammation in the mammary fat at a distance from the tumor lesion in patients (n = 9). Scale bar, 100 μm for trichrome staining and 50 μm for Col and Mac2 staining.
(I) Average fibrous area and thickness in distant and adjacent regions from the tumor lesion in patients (n = 9). Each dot represents an individual sample. Data are presented as mean ± SEM. Unpaired Student’s t test was used in (F), (G), and (I). *p < 0.05; **p < 0.01; ***p < 0.001.
