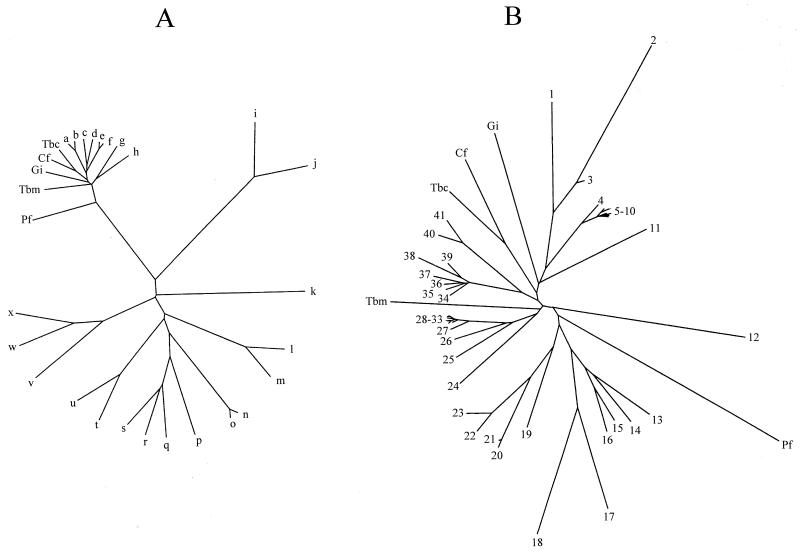
FIG. 2.
Phylogenetic analysis of parasite AspATs. In both trees, constructed by neighbor joining with the Phylip package, parasite AspATs are abbreviated as follows: Cf, C. fasciculata cytoplasmic AspAT; Tbc, T. brucei brucei cytoplasmic AspAT; Tbm, T. brucei brucei mitochondrial AspAT; Gi, G. intestinalis cytoplasmic AspAT; and Pf, P. falciparum AspAT. Tree A was formed using the following additional sequences: a, H. sapiens cytoplasmic AspAT; b, G. gallus cytoplasmic AspAT; c, Lupinus angustifolicus mitochondrial AspAT; d, Oryza sativa cytoplasmic AspAT; e, H. sapiens mitochondrial AspAT; f, G. gallus mitochondrial AspAT; g, E. coli AspAT; h, E. coli TyrAT; i, Schizosaccharomyces pombe TyrAT; j, S. cerevisiae TyrAT; k, E. coli alanine-valine aminotransferase; l, H. sapiens AlaAT; m, S. cerevisiae AlaAT; n, H. sapiens kynurenine aminotransferase; o, Rattus norvegicus kynurenine aminotransferase; p, B. subtilis YHDR gene product; q, B. subtilis AspAT; r, B. subtilis PATA gene product; s, B. subtilis YUGH gene product; t, H. sapiens TyrAT; u, Trypanosoma cruzi TyrAT; v, Halobacter sp. histidinol-phosphate aminotransferase; w, E. coli histidinol-phosphate aminotransferase; and x, S. pombe histidinol-phosphate aminotransferase. Tree B was formed using the following additional sequences from members of the aminotransferase Ia subfamily: 1, C. elegans CE07462 gene product; 2, C. elegans CE06829 gene product; 3, C. elegans CE07461 gene product; 4, G. gallus cytoplasmic AspAT; 5, Mus muris cytoplasmic AspAT; 6, R. norvegicus cytoplasmic AspAT; 7, H. sapiens cytoplasmic AspAT; 8, Equus caballus cytoplasmic AspAT; 9, Bos taurus cytoplasmic AspAT; 10, Sus scrofus cytoplasmic AspAT; 11, S. cerevisiae cytoplasmic AspAT; 12, S. cerevisiae mitochondrial AspAT; 13, Vibrio cholerae AspAT; 14, E. coli AspAT; 15, Haemophilus influenzae AspAT; 16, Neisseria gonorrhoeae AspAT; 17, Paracoccus denitrificans TyrAT; 18, Rhizobium meliloti TyrAT; 19, Pseudomonas aeruginosa TyrAT; 20, N. gonorrhoeae TyrAT; 21, N. meningitidis TyrAT; 22, E. coli TyrAT; 23, K. pneumoniae TyrAT; 24, A. thaliana AspAT1; 25, Drosophila melanogaster CT10757 gene product; 26, C. elegans CE02477 gene product; 27, G. gallus mitochondrial AspAT; 28, H. sapiens mitochondrial AspAT; 29, E. caballus mitochondrial AspAT; 30, B. taurus mitochondrial AspAT; 31, S. scrofus mitochondrial AspAT; 32, R. norvegicus mitochondrial AspAT; 33, M. muris mitochondrial AspAT; 34, Medicago sativa cytoplasmic AspAT; 35, Daucus caroti cytoplasmic AspAT; 36, O. sativa cytoplasmic AspAT; 37, A. thaliana AspAT3; 38, A. thaliana AspAT4; 39, A. thaliana AspAT2; 40, L. angustifolicus mitochondrial AspAT; and 41, A. thaliana mitochondrial AspAT.