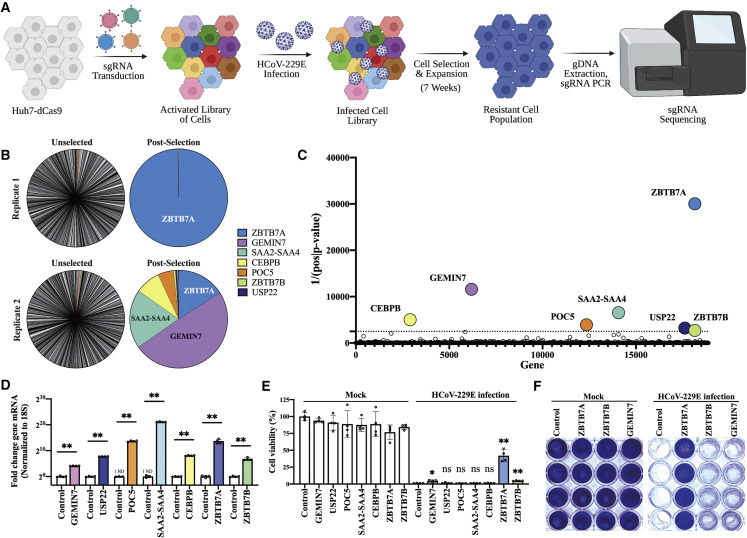
Figure 1.
CRISPR activation screen identifies host factors that allow avoidance of HCoV-229E-induced cell death
(A) Overview of CRISPR activation screen.
(B) Distribution of sgRNA reads before (unselected) and after (post-selected) infection; the most abundant 500 sgRNAs after selection are shown.
(C) The top seven enriched genes based on the inverse of the p value.
(D) Quantification of mRNA by qRT-PCR from the indicated overexpression cell lines. n = 4 biological replicates. ND, not detected.
(E) Cell viability of transduced Huh7 cells after viral infection. Transduced cells were infected with HCoV-229E at 0.5 MOI. MTT assay was conducted 3 DPI. n = 4 biological replicates.
(F) Crystal violet staining of transduced Huh7 cells after infection. Control Huh7 cells expressing mCherry, ZBTB7A-Huh7 cells, ZBTB7B-Huh7 cells, and GEMIN7-Huh7 cells were infected with HCoV-229E at 0.5 MOI. The cells were stained at 5 DPI. n = 4 biological replicates.
All panels except (A)–(C) are representative of two independent experiments. For (D) and (E), p values were calculated by unpaired two-tailed Student’s t tests. ∗p < 0.05; ∗∗p < 0.001; ns, not significant. Data shown as mean ± SD.