Fig. 4.
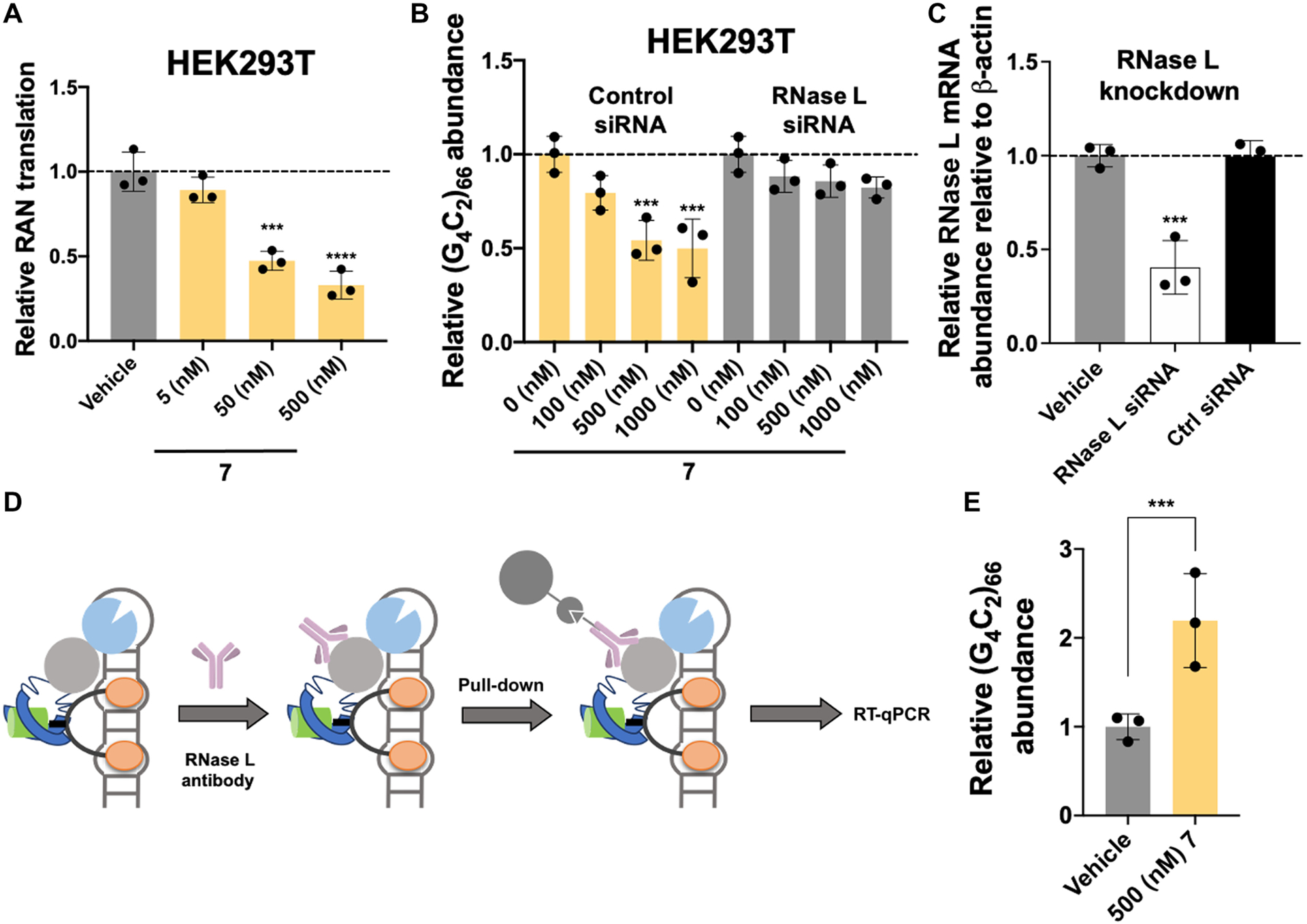
RIBOTAC 7 degrades r(G4C2)exp in a cellular model of c9ALS/FTD by local recruitment of RNase L.
(A) Effect of 7 on RAN translation, as assessed in HEK293T cells dually transfected with (G4C2)66-No ATG-Nano-Luciferase (disease) and SV40-Firefly (normalization) (n = 3 replicates). (B) Effect of 7 on r(G4C2)exp in HEK293T cells transfected with a plasmid encoding (G4C2)66-No ATG-GFP (n = 3 biological replicates) and ablation of this effect upon RNase L knockdown with an RNase L–targeting siRNA (n = 3 replicates). (C) Knockdown of RNase L by siRNA in HEK293T cells (n = 3 replicates). (D) Scheme of an RNase L immunoprecipitation (IP) assay to study ternary complex formation between r(G4C2)66, RNase L, and 7. (E) Results of the RNase L IP assay, used to assess ternary complex formation in cells (n = 3 replicates). ***P < 0.001; ****P < 0.0001, as determined by a one-way ANOVA with multiple comparisons (A and C) or unpaired t test with Welch’s correction (E). Error bars indicate SD.
