Figure 2. The cardiac enhancer landscape is highly reproducible across individuals.
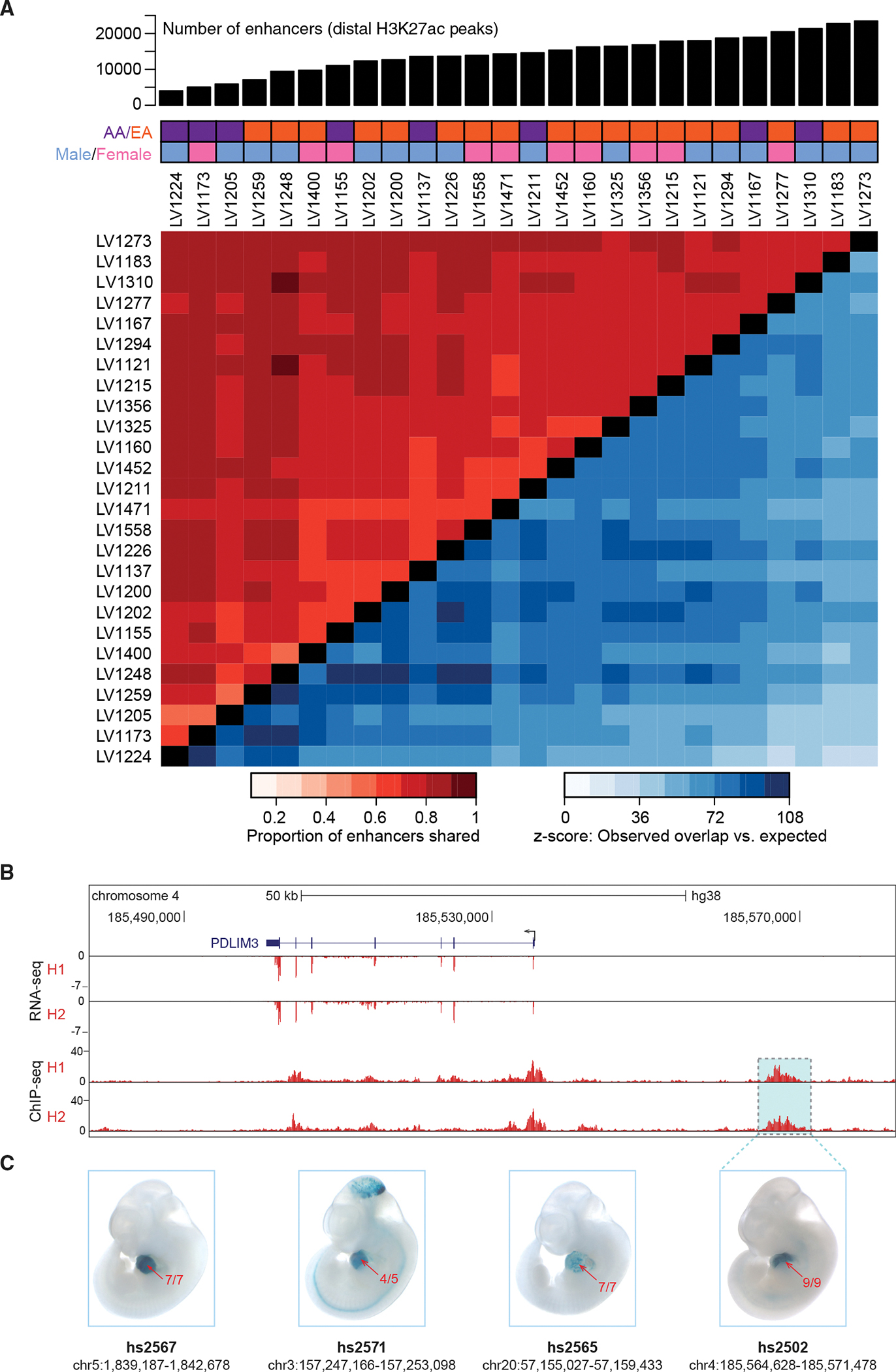
(A) Top: number of predicted enhancers per sample for 26 healthy left ventricle samples. Middle: demographic information for each subject (AA, African American; EA, European American). Bottom: Heatmap showing the proportion of peaks shared between samples (red tones in top left), along with z-scores indicating how many standard deviations the observed number of shared peaks exceeded random expectation (blue tones in bottom right, see STAR Methods).
(B) Paired RNA-seq and ChIP-seq tracks from two samples, H1 and H2, at the PDLIM3 locus.
(C) Transgenic mouse assay validation of four heart enhancers, including one upstream of PDLIM3 (see Figure S2 for results for additional predicted heart enhancers). One representative embryonic day 11.5 embryo is shown for each enhancer, and numbers in red show the reproducibility of heart staining in each transgenic experiment. Information underneath each embryo includes the identification number in the VISTA Enhancer Browser (Visel et al., 2007) (enhancer.lbl. gov) and the human genome coordinates (hg38) of each tested enhancer. Embryos have an average crown-rump length of 6 mm.
