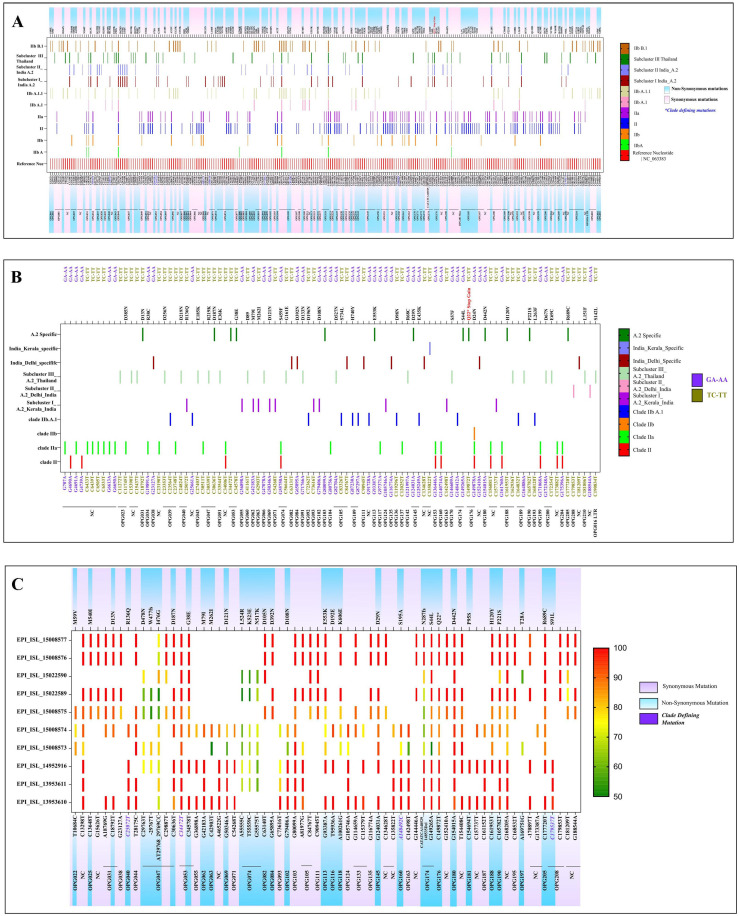
Fig. 2.
(A) The variant analysis of MPXV genome was done using software MEGA 10 w.r.t reference sequence NC_063383. The depicted figure showed the synonymous and non-synonymous mutations in Clade II and lineages. The A.2 lineage is further characterized into sub-cluster I, II and III. All the clade defining mutations are marked with blue color positions. (B) The APOBEC 3 mutation analysis was done using software MEGA 10.0. The mutations (GA-AA, TC-TT) were marked for all the clades. (C) The variant analysis of MPXV genome of all the retrieved sequences was done using CLC genomics workbench with 50% frequency. The reference positions marked with synonymous and non-synonymous mutations for all the sequences with the respective frequencies.