Figure 2.
Sparse kNN-based ATAC-RNA cell pairing allows optimal pairing and integration of scATAC-seq and scRNA-seq data
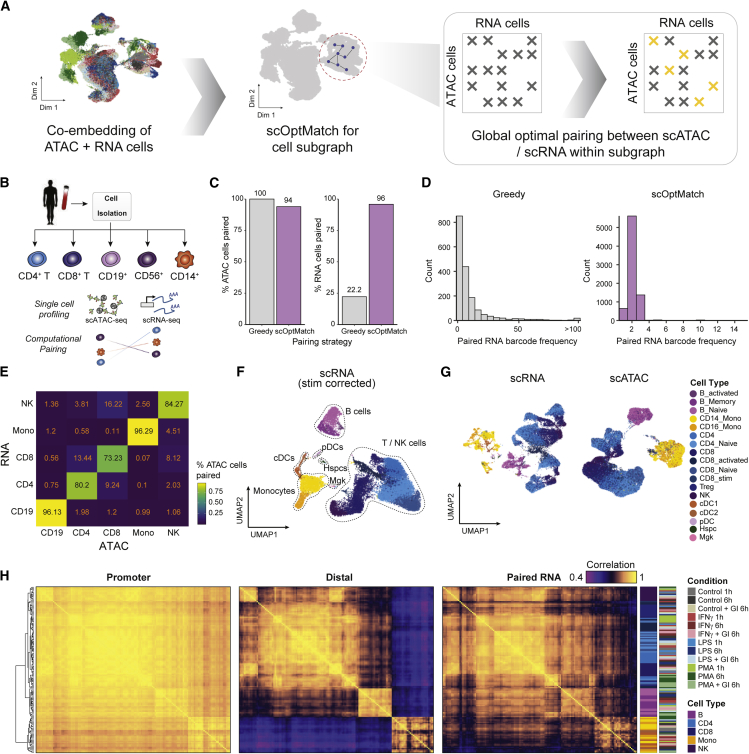
(A) Schematic highlighting scOptMatch’s strategy for computational pairing of scATAC-seq and scRNA-seq cells based on geodesic distance kNNs (yellow x marks) within cluster subgraphs (gray x marks).
(B) Schematic depicting experimental bead enrichment of specific immune cell types from human PBMCs.
(C) Distribution of the number of instances of paired RNA cell barcode when using the greedy (left) versus scOptMatch method for the PBMC isolate dataset pairing.
(D) Percentage of total scATAC and scRNA-seq cells paired using the two different pairing strategies.
(E) Accuracy heatmap of scATAC-scRNA-seq pairing between PBMC isolate cell types, colored by percentage of scATAC-seq cells correctly paired with the corresponding scRNA-seq cell type.
(F) UMAP of scRNA-seq stimulated cells shown in Figure 1D, with cells aligned across stimulus conditions to enable cell type annotation, colored by annotated cell type.
(G) UMAP of un-aligned scRNA-seq cells (shown in Figure 1D) colored by annotated cell type (left) and scATAC-seq stimulated cells (shown in Figure 1C) colored by paired scRNA-seq cell annotations (right), enabling downstream data integration for stimulated scATAC- and scRNA-seq-profiled cells.
(H) Pairwise Pearson correlation of aggregate single-cell chromatin accessibility profiles associated with gene promoters (left), distal from the promoter (center) and paired gene expression (right), aggregated by cell type and condition.