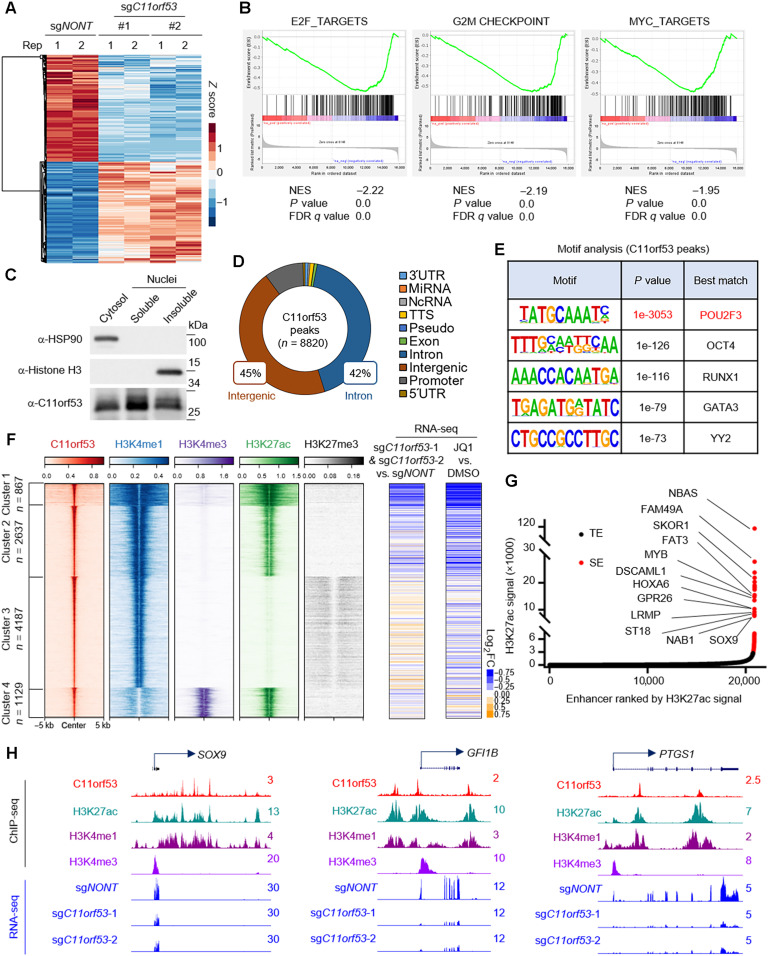
Fig. 3. C11orf53 regulates lineage-specific gene expression at super enhancers.
(A) RNA-seq was conducted with NCI-H526 cell lines transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs for 4 days. The heatmap shows the differentially regulated genes, n = 2. (B) The GSEA plot shows the enrichment of E2F, G2M checkpoint, and MYC pathways genes enriched in the down-regulated genes with C11orf53 depletion. NES, normalized enrichment score; FDR, false discovery rate. (C) The protein levels of C11orf53 in cytosol, soluble nuclear fraction, and insoluble nuclear fraction was determined by Western blot. HSP90 was used as cytoplasmic protein control, and the histone H3 was used as nuclear insoluble protein control. (D) The pie plot shows the annotation and distribution of C11orf53 peaks at the genome. (E) Motif analysis shows the best-matched motifs occupied by C11orf53. (F) The total C11orf53 peaks were divided into four clusters based on k-means clustering. The histone marks were further centered on C11orf53 peaks in each cluster (left). RNA-seq was conducted with cells transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs. The log2 fold-change (FC) heatmaps shows the expression change of nearest genes to C11orf53 peaks (middle), n = 2. RNA-seq was conducted with cells treated with either dimethyl sulfoxide (DMSO) or JQ1 (1 μM). The log2 fold-change heatmaps shows the expression change of nearest genes to C11ORF53 peaks (right), n = 2. (G) The Histone H3 lysine 27 acetylation (H3K27ac) signals from chromatin immunoprecipitation (ChIP) sequencing identifies putative super enhancers (SEs) in NCI-H526 cells. Hockey-stick plot representing the normalized rank and signals of H3K27ac. Representative SE-associated genes that are occupied by C11orf53 are labeled. TE, typical enhancers. (H) Representative tracks showing the enhancer binding of C11orf53, which contributes to activation of gene expression.