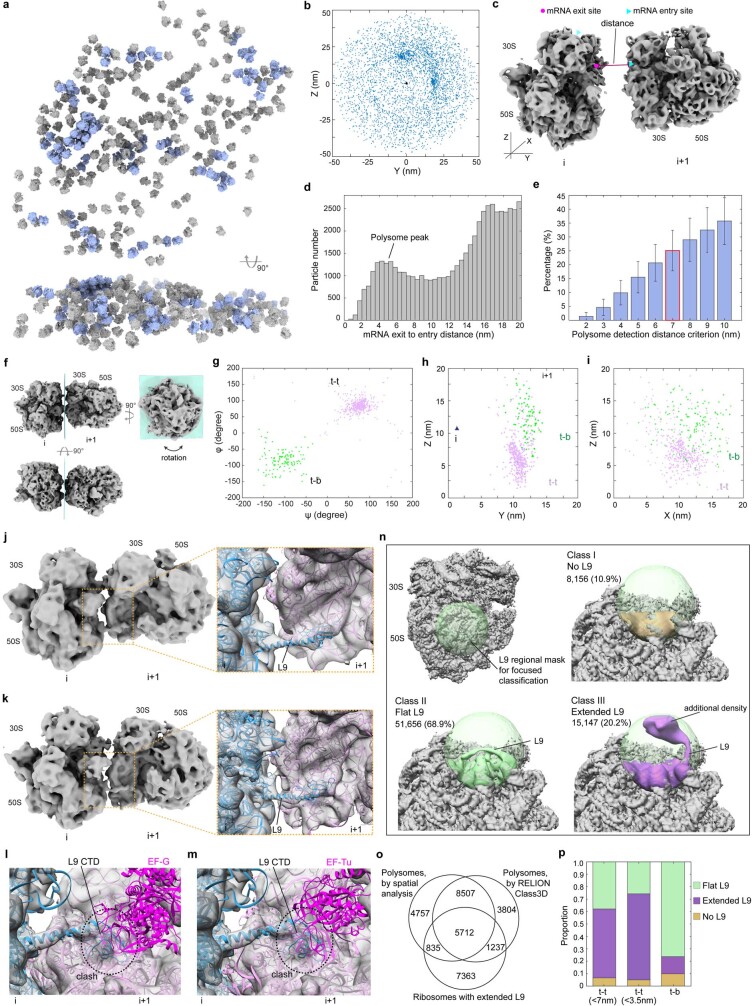
Extended Data Fig. 12. Spatial analysis of ribosomes and polysomes in native untreated cells.
a, 70S ribosomes (grey) and detected polysomes (light blue) in a representative tomogram. b, Distribution of neighbouring ribosomes within a 50 nm centre-to-centre distance. Rotation normalized. c, Illustration of the polysome detection approach based on the distance from the mRNA exit site of one ribosome to the mRNA entry site of the next. d, Histogram of distances from the mRNA exit site of one ribosome to the mRNA entry sites of all neighbouring ribosomes. e, Percentages of ribosomes detected as polysomes using different distance thresholds (d). Mean and s.d. are across 356 tomograms (n = 356 cells). A 7 nm threshold was selected. f, In polysomes, the following ribosome (i) adopts various orientations relative to the preceding ribosome (i+1), mainly with regard to rotation around a plane perpendicular to the preceding ribosome’s mRNA exit site. g, Rotations of the following ribosome relative to the preceding ribosome in polysomes. The two clusters correspond to the previously defined 't-t' and 't-b' configurations. h-i, Positions of the following ribosomes (coloured according to different rotations) relative to the preceding ribosome (triangle indicates the mRNA exit site). j-k, Two representative di-ribosome pairs in polysomes with extended L9. l-m, Clash between the C-terminal domain of the extended L9 and the superposed EF-G and EF-Tu, respectively. n, De novo focused classification on the L9 region using a spherical mask (light green) of 70S ribosomes resulted in three classes: no L9 resolved (I), flat L9 (II) and extended L9 (III). The additional density connecting to the extended L9 originates from the neighbouring ribosome. o, Overlap between polysomes independently defined based on spatial analysis, RELION classification for polysomes and classified ribosomes with the extended L9. p, The extended L9 is more frequent in the 't-t' ribosome pairs, especially in compacted ones.