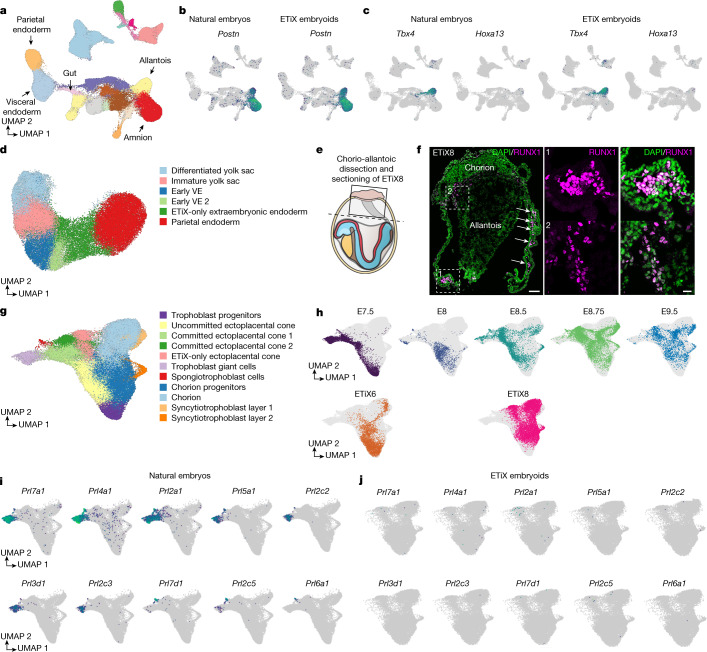
Fig. 6. Characterization of extraembryonic lineages in ETiX embryoids.
a, Global UMAP of the tiny-sci dataset as shown in Fig. 1f; selected cell clusters are highlighted. b, Gene expression of the amnion marker periostin (Postn) in natural embryos and ETiX embryoids from the tiny-sci-RNA-seq dataset. c, Gene expression of the allantois markers Tbx4 and Hoxa13 in natural embryos and ETiX embryoids from the tiny-sci-RNA-seq dataset. d, Subclustered and annotated UMAP of extraembryonic endoderm from the tiny-sci-RNA-seq dataset. e, Schematic of dissection of chorioallantoic attachment of ETiX embryoids. f, Left, sagittal section of chorioallantoic attachment and yolk sac of day 8 ETiX embryoid showing RUNX1 and DNA. Arrows highlight blood islands. Outlined regions are magnified in the middle and right panels. n = 3 ETiX8 from 3 experiments. Scale bars: 100 μm (left), 20 μm (middle and right). g, Subclustered and annotated UMAP of ExE and trophoblast cells from the tiny-sci-RNA-seq dataset. h, The contribution of individual timepoints to the subclustered UMAP of the ExE and trophoblast cells. i,j, Expression of selected prolactin genes in the subclustered UMAP of ExE and trophoblast cells for natural embryos (i) and ETiX embryoids (j).