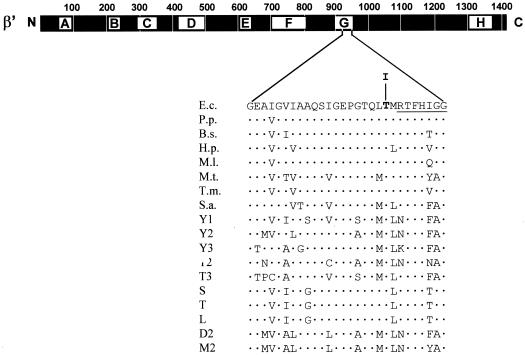
FIG. 2.
Genetic context and sequence alteration of the rpoC931 mutation. The heavy bar represents the 1,407-amino-acid β′ subunit of E. coli RNAP. Lettered boxes (A to H) symbolize evolutionarily conserved segments of β′ (33). A relevant sequence stretch (Gly912 to Gly939) from segment G of β′ (E.c.) is expanded underneath. The position of the amino acid alteration (a Thr-to-Ile change at codon 931) caused by the rpoC931 mutation is highlighted by boldface. The top sequence from E. coli is aligned with the corresponding portions of eubacterial, archaeal, and eukaryotic homologues: Pseudomonas putida (P.p.); Bacillus subtilis (B.s.); Helicobacter pylori (H.p.); Mycobacterium leprae (M.l.); Methanobacterium thermoautotrophicum (M.t.); Thermotoga maritima (T.m.); Sulfolobus acidocaldarium (S.a.); Saccharomyces cerevisiae RNAP I, II, and III (Y1, Y2, and Y3, respectively); RNAP II and III of Trypanosoma (T2 and T3, respectively); chloroplasts from spinach (S), tobacco (T), and liverwort (L); RNAP II of Drosophila melanogaster (D2); and RNAP II of mouse (M2). The dots symbolize amino acid sequence identity. The highly conserved 7-amino-acid sequence stretch (RTFHIGG) shown by Borukhov et al. (4) to contact the nascent RNA product is underlined.