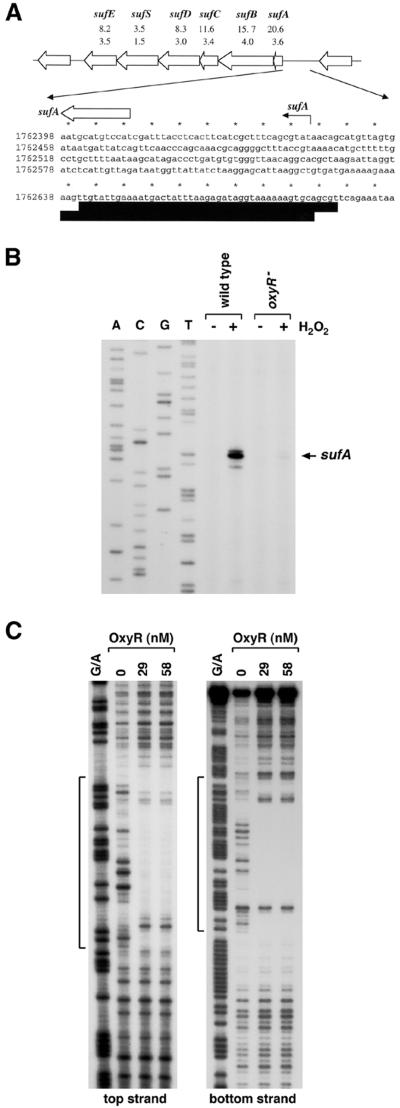
FIG. 2.
OxyR-dependent induction of the suf operon. (A) Structure of the suf operon and sequence of the sufA promoter. The induction ratios observed for the wild-type and ΔoxyR mutant strains in the microarray experiment are given below each gene. The sufA transcription start is marked by the black arrow, and the start of the corresponding ORF is denoted by a white arrow. The DNase I footprints for OxyR binding are indicated by the dark boxes. Our computational search (41) predicted an OxyR binding site of 9.6 bits centered at position 922026. (B) Primer extension assays of sufA expression in wild-type (MC4100) and ΔoxyR (GSO47) strains grown in LB medium. Exponential-phase cultures were split into two aliquots: one aliquot was left untreated, and the other was treated with 1 mM hydrogen peroxide. The cells were harvested after 10 min, total RNA was isolated, and primer extension assays were carried out with primer 820 specific to sufA. The neighboring sequencing reactions were carried out with the same primer. (C) DNase I footprinting assays of oxidized OxyR binding to the top and bottom strands relative to the sufA promoter. The regions protected by OxyR on both strands are indicated by the brackets. For OxyR binding to the top strand, the 32P-labeled primer 828 and unlabeled primer 825 were used to PCR amplify a 187-bp fragment. For OxyR binding to the bottom strand, the 378-bp NotI-BamHI fragment of pGSO133 was labeled with 32P at the BamHI site. The samples were run in parallel with Maxam-Gilbert G/A sequencing ladders.