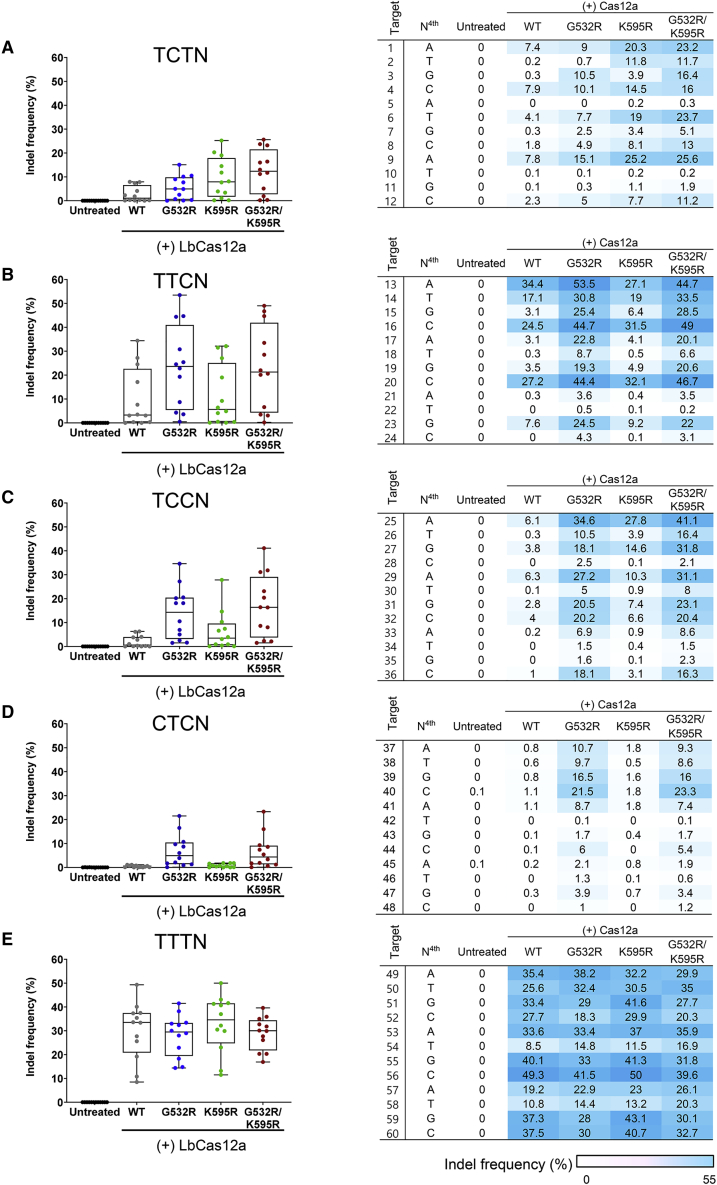
Figure 2.
Activity of engineered LbCas12a variants at sites with non-canonical and canonical PAMs
(A–E) Comparison of the genome editing activities of wild-type LbCas12a and LbCas12a variants harboring G532R, K595R, or G532R/K595R mutations measured by targeted deep sequencing at 60 endogenous target sites in HEK293T cells. Data points (n = 12) are plotted as dots representing the crRNAs indicated in Table S2. Each dot represents the mean of biologically independent triplicates. (A) TCTN, (B) TTCN, (C) TCCN, (D) CTCN, and (E) TTTN PAMs. The heatmap shows the mean indel frequencies at 60 target sites with TCTN, TTCN, TCCN, CTCN, and TTTN PAMs.