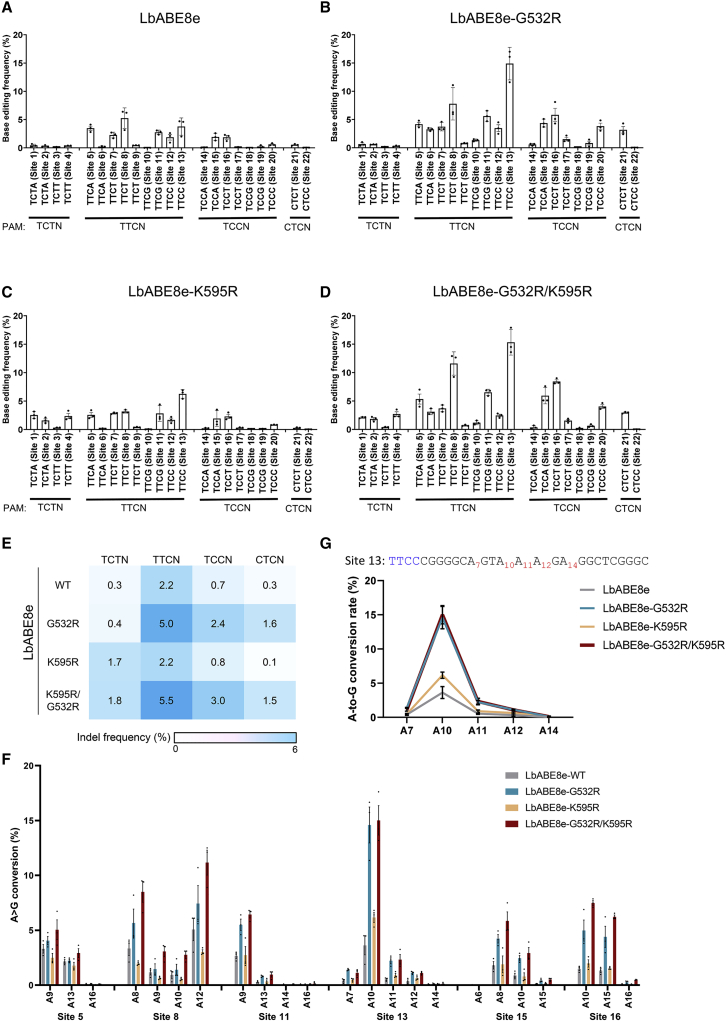
Figure 4.
Adenine base editing with engineered LbABE8e variants at sites with non-canonical PAMs
Adenine base editing efficiencies induced by (A) wild-type LbABE8e and engineered LbABE8e containing (B) G532R, (C) K595R, or (D) G532R/K595R mutations at 22 endogenous target sites, at which indels were generated by LbCas12a-G532R/K595R at frequencies above 20%, measured by targeted deep sequencing in HEK293T cells. Mean base editing frequencies ± SEM are shown in the graphs. (E) A representative heatmap shows mean base editing frequencies measured by targeted deep sequencing. (F) The base editing window within the protospacer at six genomic sites in HEK293T cells for wild-type LbABE8e and engineered LbABE8e containing G532R, K595R, or G532R/K595R mutations. (G) The enhanced editing window of LbABE8e variants at site 13. Each dot represents an individual data point. Error bars indicate SEM (n = 3).