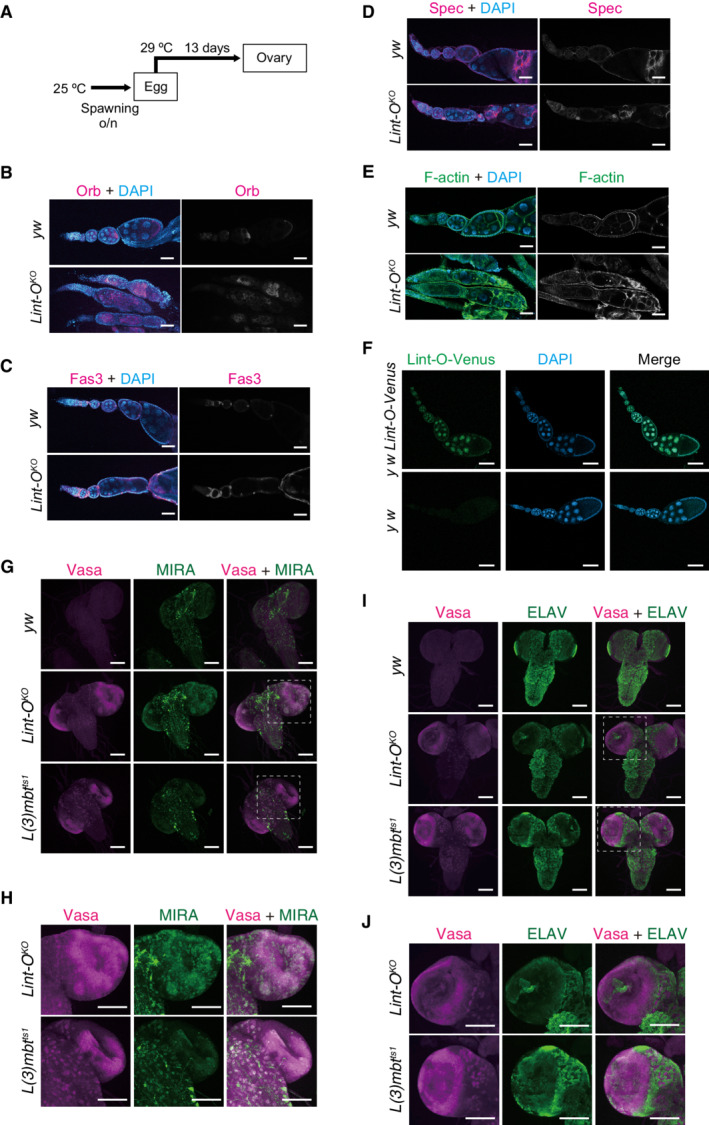
Figure EV5. Lint‐O KO ovaries and larval brains .
-
ASchema depicting the experiment. Eggs were transferred to 29°C after spawning at 25°C and incubated for 13 days. Ovaries were dissected from adult flies.
-
B–EConfocal images of brains of y w, Lint‐O KO , and L(3)mbt ts1 larvae (grown at 29°C) for Orb (B), Fas3 (C), Spectrin (Spec) (D), and F‐actin (E). Lint‐O KO ovariole showed defects in follicle cell layer integrity. DAPI (blue): nuclei. Scale bars: 50 μm.
-
FThe Lint‐O‐Venus fly line (y w Lint‐O‐Venus) was generated using the CRISPR /Cas9 system. The Venus sequence was inserted before the stop codon of Lint‐O. The Lint‐O‐Venus signal (green) was detected in the nucleus of both germ and follicle cells in the ovaries. Nuclei were stained with DAPI (blue). Wild‐type (y w) was used as a negative control. Scale bars: 50 μm.
-
GConfocal images of y w, Lint‐O KO , and L(3)mbt ts1 immunostained for Vasa (magenta) and MIRA (green). Merged images were also presented in Fig 6B. Scale bars: 100 μm.
-
HEnlarged views of insets in (G). Scale bars: 50 μm.
-
IConfocal images of y w, Lint‐O KO , and L(3)mbt ts1 immunostained for Vasa (magenta) and ELAV (green). Merged images were also presented in Fig 6B. Scale bars: 100 μm.
-
JEnlarged views of insets in (I). Scale bars: 100 μm.
