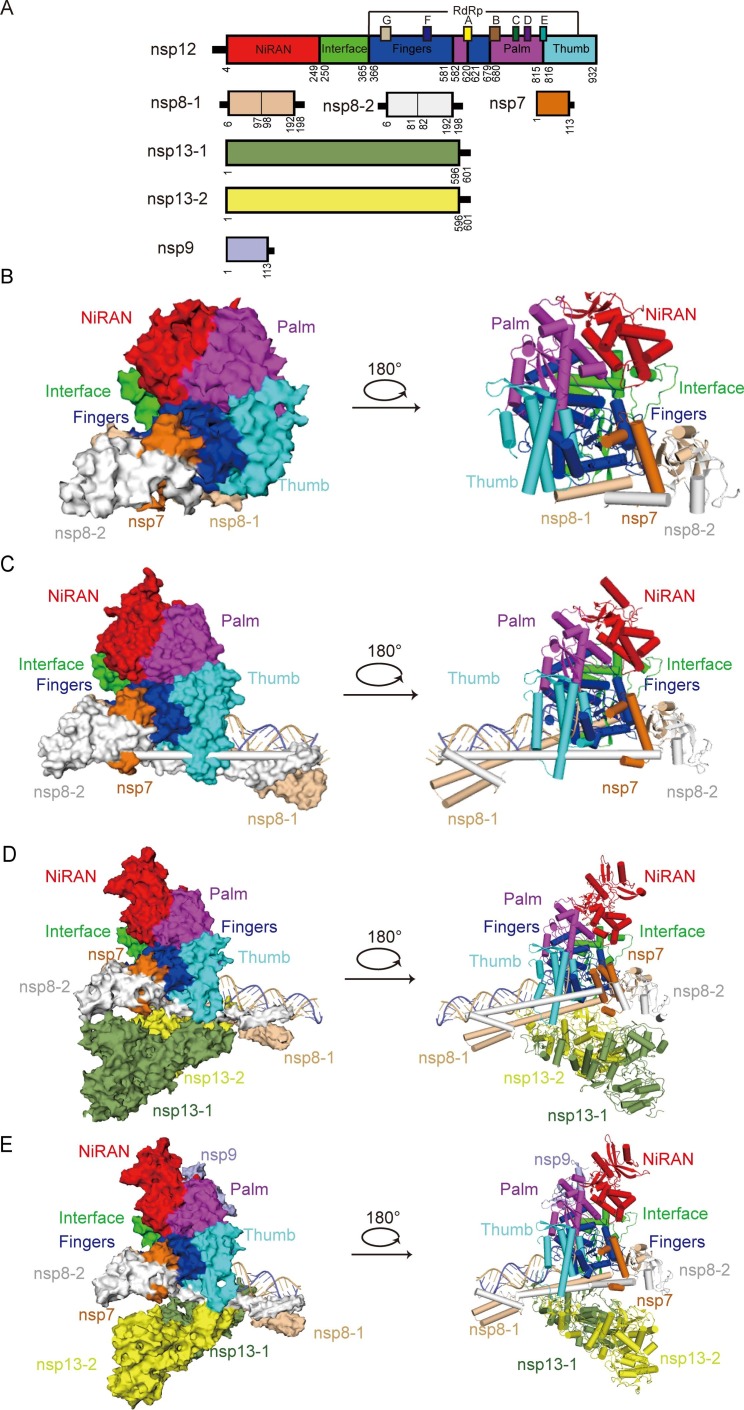
Fig. 2.
Structure of SARS-CoV-2 replication-transcription complex (RTC) (A) The schematic diagram for the domain organization of each component in the RTC, containing nsp12, nsp7, two copies of nsp8 (nsp8-1 and nsp8-2), two copies of nsp13 (nsp13-1 and nsp13-2) and nsp9. The interdomain borders are labeled with residue numbers. In nsp12, the conserved sequence motifs A-G are depicted. The components and domains are colored as follows: nsp12 NiRAN, red; nsp12 Interface, green; nsp12 Fingers, blue; nsp12 Palm, magenta; nsp12 Thumb, cyan; nsp7, orange; nsp8-1, wheat; nsp8-2, white; nsp13-1, split-pea; nsp13-2, yellow; nsp9, light blue. The color coding corresponds to the figures throughout this manuscript. (B) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 apo RdRp complex (nsp7-nsp82-nsp12) in different views (PDB ID:7BW4). (C) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 polymerase in complexes with RNA (nsp7-nsp82-nsp12-RNA, central RTC) in different views (PDB ID:6YYT). The RNA is depicted with the template strand in slate and the primer/product strand in light orange. (D) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 central RTC with nsp13 helicases (nsp7-nsp82-nsp12-nsp132-RNA, elongation RTC) in different views (PDB ID:6XEZ). (E) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 elongation RTC with nsp9 (nsp7-nsp82-nsp12-nsp132-nsp9-RNA, extended E-RTC) in different views (PDB ID:7CYQ). Nsp9 binds to the catalytic center of the nsp12 (RdRp) NiRAN domain.