Abstract
Pregnancy success requires constant dialogue between the mother and developing conceptus. Such crosstalk is facilitated through complex interactions between maternal and fetal cells at distinct tissue sites, collectively termed the maternal-fetal interface. The emergence of single-cell technologies has enabled a deeper understanding of the distinct processes taking place at the maternal-fetal interface as well as the discovery of novel pathways and immune and non-immune cell types. Single-cell approaches have also been applied to decipher the cellular dynamics throughout pregnancy, in parturition, and in obstetrical syndromes such as recurrent spontaneous abortion, preeclampsia, and preterm labor. Furthermore, single-cell technologies have been utilized during the recent COVID-19 pandemic to evaluate placental viral cell entry and the impact of SARS-CoV-2 infection on maternal and fetal immunity. In this review, we summarize the current knowledge of cellular immunobiology in pregnancy and its complications that has been generated through single-cell investigations of the maternal-fetal interface.
Introduction
Pregnancy is a dynamic period of growth and development that is initiated at fecundation and culminates in parturition, resulting in the delivery of the conceptus (1). To successfully complete this journey, a coordinated series of complex events is required, which starts with the establishment of dialogue between the mother and embryo (2–9). Such communication is facilitated through multiple sites of maternal-fetal interaction, which have been collectively termed the maternal-fetal interface (10–15). Such sites represent anatomically distinct areas of immunological contact; namely, the decidua basalis, where maternal immune cells and decidual stromal cells interact with fetal extravillous trophoblast; the placental intervillous space, in which circulating maternal immune cells interact with the fetal syncytiotrophoblast; and the boundary where the decidua parietalis meets the chorion laeve in the chorioamniotic membranes (11, 13, 14, 16). Therefore, the maternal-fetal interface comprises numerous complex immune and non-immune cellular interactions that support implantation (17–21), promote fetal and placental development (22, 23), maintain homeostasis (17, 19, 24–37), and participate in the inflammatory milieu implicated in parturition (38–47). Accordingly, the disruption of maternal-fetal homeostasis at different stages of pregnancy has been implicated in multiple pathologies ranging from pregnancy loss to preterm delivery (26, 27, 30–32, 36, 37, 44, 45, 48–51). Hence, deciphering the mechanisms of maternal-fetal crosstalk that lead to a normal term delivery as well as those implicated in obstetrical disease is paramount.
Recent investigations have leveraged the use of single-cell technologies to evaluate the transcriptome, proteome, and epigenome of the maternal-fetal interface at single-cell resolution. Among these, single-cell RNA-sequencing (scRNA-seq) was the first to be described (52–60) and has seen the most widespread use across reproductive science (61–65). Indeed, scRNA-seq was utilized to provide the first insights into the cellular heterogeneity and interaction networks of the maternal-fetal interface in early (66–77) and late (78–80) gestation, and the resulting datasets of these pioneer studies have since been integrated into a number of subsequent investigations. Furthermore, single-cell technologies have been highly effective at uncovering the unique biological processes taking place at the maternal-fetal unit during mid-gestation (66, 81–85) as well as those inflammatory pathways implicated in the processes of term and preterm parturition (80, 86–90). Pertinent to the current pandemic, scRNA-seq has also provided important knowledge about the mechanisms of cell entry for SARS-CoV-2 as well as its deleterious effects on the maternal-fetal unit (91–99). Herein, we aim to provide an overview of such studies in a succinct and comprehensive manner to foster future research utilizing single-cell technologies to decipher the cellular mechanisms of disease for pregnancy complications.
The first trimester
The first trimester represents an important developmental window for the fetus, which relies heavily on proper placentation and spiral artery remodeling (100–104). To aid in these processes, a carefully orchestrated series of interactions is required that involves maternal pro-angiogenic or tolerogenic immune cells, mesenchymal, endothelial, and decidual stromal cells, and the invading fetal trophoblast. The first trimester has therefore been an attractive target period for investigating the cellular states, functions, and interactions that drive pregnancy establishment (66–77). Moreover, due to the importance of a healthy first trimester for continued pregnancy, multiple single-cell studies have targeted the maternal-fetal interface during this time to help unravel the mechanisms underlying pathologies such as recurrent spontaneous abortion (71, 73–75, 77) (Figure 1). Next, we summarize the insights into the maternal-fetal immunobiology of the first trimester obtained using single-cell technologies.
Figure 1. The single-cell immunobiology of the maternal-fetal interface in early pregnancy.
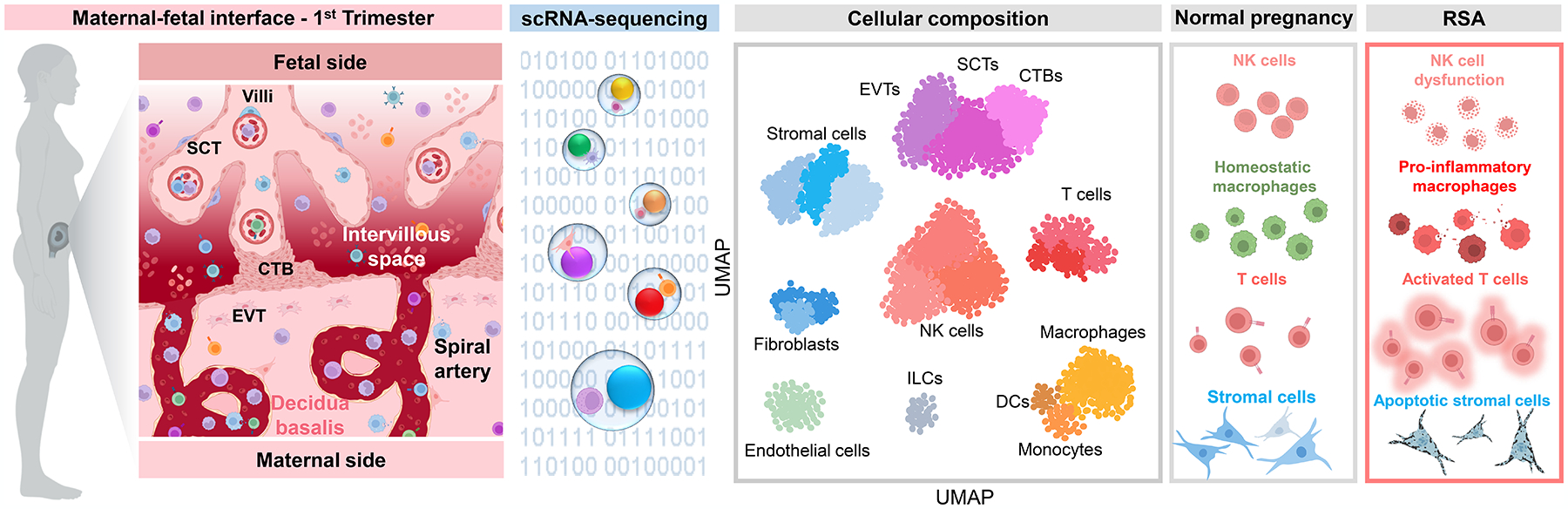
The first-trimester maternal-fetal interface is marked by placental growth, trophoblast invasion and angiogenesis, and the establishment of maternal-fetal dialogue to promote homeostasis for the remainder of pregnancy. During this period, the placental compartment primarily comprises trophoblast cell types, fibroblasts, stromal cells, and Hofbauer cells (fetal macrophages), as represented in the UMAP plot based on data reported by Vento-Tormo et al. 2018. By contrast, the decidua displays more heterogeneous cellularity, characterized by major populations of decidual stromal cells, NK cells, macrophages, and endothelial cells. Other leukocyte subsets are also represented in the early decidua; namely, T cells, innate lymphoid cells, and dendritic cells. Single-cell investigations of recurrent spontaneous abortion (RSA) have suggested that this disease includes the altered composition of decidual NK cells and generalized T-cell infiltration and activation, together with the acquisition of a pro-inflammatory phenotype by NK cells and macrophages. Indeed, it was proposed that a shift away from macrophage-NK cell interactions toward macrophage-T cell interactions may contribute to RSA. The decidual stromal compartment may also be affected, as enrichment of cellular processes related to apoptosis and senescence implicates impaired stromal cell differentiation in RSA pathophysiology. Abbreviations used: SCT, syncytiotrophoblast; CTB, cytotrophoblast; EVT, extravillous trophoblast; NK cell, natural killer cell; DC, dendritic cell; ILC, innate lymphoid cell; UMAP, uniform manifold approximation and projection. Figure created using BioRender.
Placental cells in the first trimester
Some of the first studies of the early placenta utilized scRNA-seq to characterize the major cell types and their subsets present in this organ (66–68). One survey utilized two scRNA-seq platforms (commercial 10x Genomics platform and a custom Drop-seq platform) to characterize the placental villi, and found good correlation between these technologies (67). Two other investigations of the first trimester maternal-fetal interface utilized either Smart-seq2 alone (66) or an integrated dataset obtained using Smart-seq2 and 10x Genomics (68). Placental cell types were largely consistent between studies, with villous cytotrophoblasts (VCTs), syncytiotrophoblasts (SCTs, also referred to as STBs), extravillous trophoblasts (EVTs), fibroblasts, endothelial cells, erythroblasts, and Hofbauer cells all being identified by their expression profiles (66–69). Comparison of relative abundance indicated that, after trophoblast subsets, the most represented placental cell type was fibroblasts, followed by endothelial cells, erythroblasts, and Hofbauer cells (67), highlighting the capacity of scRNA-seq to map the cellular heterogeneity of the placenta.
In addition to their identification, the further characterization of trophoblast cell types revealed that CTBs and EVTs include multiple subsets that displayed differing fusion potential or proliferative capacity, respectively (66). One of these CTB subsets displayed comparatively higher Syncytin-2 expression (66), suggesting fusion competence that would allow SCT formation (105). Pseudotime analysis indicated a transition from CTB to EVT subtypes (66), consistent with other reports that proposed two trophoblast differentiation pathways by which VCT could transition to the EVT or SCT subsets (68). Such endpoint EVTs represented the most invasive trophoblast subset that is found in the maternal decidua (68) and characterized by expression of specific factors such as SOX4 (70). A recent investigation utilized scRNA-seq of the first trimester placenta together with a prior dataset (68) to assess the trophoblast cell states implicated in the establishment, renewal, and differentiation of these cells (76). Four CTB states (CTB1–4), a SCT precursor-like CTB state (SCTp), two column trophoblast (EVT precursor (106)) states, and an EVT state were identified based on gene expression (76). While the relative frequencies of the four CTB states increased throughout the first trimester, the column trophoblast and EVT states decreased from early to late first trimester and SCT progenitors remained constant (76), highlighting the fluctuation of trophoblast cell states during this period. Combined RNA velocity and single-cell trajectory analyses were used to map primitive trophoblasts to the CTB2/CTB3 clusters, from which two putative differentiation pathways emerged: one directed toward EVT and the other toward SCTp (76). To validate this finding, Monocle 3 and Slingshot were applied and agreement between these two pseudotime analyses was demonstrated, indicating CTB2 as the closest point of origin (76). Thus, scRNA-seq allows for the dissection of the origins and differentiation trajectories of the heterogeneous cell types that form the placenta.
It is worth mentioning the tools that have been developed for trajectory inference by which single cells are ordered based on similarities in gene expression (107), as was performed in the studies described above. A large number of trajectory inference methods have been developed, with more being generated often; yet, the largest difference among them is whether the topology is fixed and, if not, what kind of topology can be detected (107). A thorough comparison of current trajectory inference techniques indicated high variability between methods, and thus it is clear that investigators should test multiple methods with their data, and more so when a priori knowledge of topology is lacking (107). Importantly, the effort required to identify and utilize such tools has been greatly diminished by the availability of resources and databases that can guide users to the best trajectory inference approaches (107–109).
The characterization of other major placental cell types revealed that, in addition to trophoblasts, the fibroblast population includes multiple subsets, with two of these displaying expression profiles reminiscent of myofibroblasts (67), which could support the contractile properties of anchoring villi (110). Hofbauer cells were also found to include two subsets, of which one was found to express an activated, MHC-II-expressing state that could be important for debris clearance in the developing placenta (66). Other cell types identified in the placenta include two distinct subsets of mesenchymal stromal cell (MSC), which displayed gene expression profiles indicative of participation in cell adhesion/migration or angiogenic processes, respectively (66).
The abovementioned reports show that scRNA-seq represents a useful technology for uncovering the composition, differentiation pathways, and expression profiles of cell types in the placenta, a unique organ that carries out critical functions for fetal growth and development (111, 112). Importantly, such knowledge can be leveraged to unravel the molecular underpinnings of placental diseases.
Decidual cells in the first trimester
The first trimester decidua represents a more heterogeneous cellular compartment compared to the placenta, given the increased abundance of immune cell types in addition to tissue cells (67, 68, 113). Natural killer (NK) cells, antigen-presenting cells (APCs) such as dendritic cells (DCs), macrophages, innate lymphoid cells (ILCs), and T cells represent major subsets in the decidua, in addition to decidual stromal cells (DSCs), fibroblasts, smooth muscle cells, endometrial epithelial cells, lymphatic endothelial cells, and vascular endothelial cells (67, 68, 113). The DSC compartment comprises multiple subsets characterized by their spatial location as well as their expression profiles (68, 71). Pseudotime analysis of DSCs and fibroblasts indicated that the fibroblast 1 subset can either differentiate to DSCs or fibroblast 2 (67). The differentiation of DSCs was further explored in a recent study of the murine uterus, in which the single-cell profiles of this tissue were compared between implantation sites (decidualized uterus) and inter-implantation sites (non-decidualized uterus) (72). In this study, the decidual cellular compartment was subdivided into deep stromal, proliferating deep stromal, superficial stromal, intermediate decidual, and decidual cells, each with expression profiles indicative of distinct functionality (72). Two potential pseudotime differentiation pathways were determined: the first was a deep stromal > superficial stromal transition, and the second was a deep stromal > proliferative deep stromal > intermediate decidual/decidual transition, which was considered as corresponding to decidualization (72). Consistent with prior receptor-ligand interaction analyses in the decidua (67, 68), interaction networks between decidual cells and different major subsets such as immune cells, endothelial cells, and trophoblast were also demonstrated (72). Interactions between decidual and immune cells included immunological pathways such as cytokine-cytokine receptor interaction, phagosome, natural killer cell-mediated cytotoxicity, and NOD-like receptor signaling pathway, among others (72). Interestingly, interactions between decidual and endothelial cells showed substantial overlap with those between decidual and immune cells, as did the interactions between decidual cells and trophoblasts; notable terms shared among all three interactions included the TGF-beta signaling pathway and the MAPK signaling pathway (72). Thus, decidualization involves a core set of signaling pathways that are shared among multiple cell types in this compartment, suggesting that this is a cooperative process. Expanding on this approach, a separate investigation utilized scRNA-seq data of the murine day 7.5 uterus to deconvolute spatial transcriptomics data derived from the same tissue, which allows for the assignment of single cells to specific uterine microenvironments (114). The integration of these datasets revealed the distinct distribution of different stromal subsets within the uterus as well as their clustering in relation to other cell types (114). Thus, the spatial localization of cell subsets at the maternal-fetal interface may provide insight into their functionality and potential interactions with neighboring cells.
The inference of cell-cell communications from scRNA-seq data has been an area of ongoing research. The first iteration of tools developed for this purpose was based on the expression of single receptor-ligand pairs (115–117), which did not account for receptors that function as multi-unit complexes. To overcome such a limitation, the CellPhoneDB v2.0 tool was developed, which could predict enriched cell-cell signaling based on the minimum average expression of all members of multi-unit complexes (118). More recently, this concept was further built upon by considering signaling cofactors such as soluble agonists/antagonists and membrane-bound co-receptors, which resulted in the creation of the cell-cell communication analysis tool CellChat (119). While such databases provide a powerful new means of exploring scRNA-seq datasets, it should be considered that the resulting inferences are based solely on gene expression and thus require careful interpretation.
Other key players during early human pregnancy are decidual NK cells, which can be classified into different subsets based on single-cell clustering (67, 68, 73) or imaging mass cytometry (IMC) (113). Among these, a study reported that one NK subsets (dNK1) is the most responsive to HLA-C/HLA-G and expresses more cytoplasmic granules compared to other subsets (68); thus, these decidual immune cells cells may be central for modulating EVT invasion during the first trimester. Pseudotime analysis of decidual NK cells indicated a potential differentiation pathway wherein the endpoint subsets showed enrichment of similar immune-related processes (73). However, alternative pathways wherein endpoint NK cells can return to an intermediate state have also been described (74). Decidual NK cells represent an important cellular contributor to pregnancy maintenance, given that a disruption in their functions (such as by poor maternal-fetal KIR-HLA matching) has been linked to complications such as spontaneous abortion or preeclampsia (120–123). A recent IMC-based investigation of the maternal-fetal interface identified six distinct clusters of NK cells (113). Consistent with a prior demonstration (68), a subset of CD69-expressing decidual NK cells (termed dNK1) was the most prominent in the first trimester (113). In addition to NK cells, a subset of T cells with a cytotoxic expression profile is also found in the first-trimester decidua (67), which may participate in the process of EVT invasion and remodeling. Other notable immune cells subsets at the early maternal-fetal interface include macrophages, which are reported to display single-cell gene expression profiles that are distinct from the conventional M1/M2 paradigm (75). The latter observation is consistent with the recently proposed role of decidual macrophages as homeostatic sentinels that promote fetal growth and sustain pregnancy in early (32) and late (37) gestation. Indeed, analysis of cell clustering based on IMC revealed that myeloid cells are most prominently localized to the trophoblast microenvironment as well as the microenvironment of other immune cells at the maternal-fetal interface (113). Moreover, among the six identified myeloid clusters, two were specifically enriched in the first trimester and lacked HLA-DR expression (113). These observations lend further support to the role of decidual macrophages as homeostatic mediators, even in the first trimester.
Taken together, these data provide deep insight into the cellular processes that characterize the first trimester decidua. In particular, single-cell transcriptomic analysis is useful for uncovering the interactions between critical immune cells (such as NK cells) and stromal cells, which can inform the overall cellular dynamics of the maternal-fetal interface.
Cellular interactions at the maternal-fetal interface during the first trimester
A key strength of scRNA-seq is the capacity to infer cellular interactions within the target tissue using analytical methods such as CellChat, which is based on the expression of receptors and ligands by cell type pairs (119). Mapping of the mostly highly expressed receptor-ligand pairs among placental and decidual cells revealed prevalent cell-cell interactions taking place in these compartments (67, 68). Notable cell-cell interactions at the maternal-fetal interface include those between EVTs and maternal immune cells, which comprise signaling related to immunomodulation, growth, angiogenesis, and cell adhesion/recruitment (68). Yet, complex interaction networks also occur among the DSCs, fibroblasts, endothelial cells, NK cells, and macrophages to support cellular differentiation and homeostasis (67, 68). Moreover, cell-cell interaction analyses are not confined to a single tissue, as studies incorporating both the decidua and placental villi have provided an overview of the strongest intercellular interactions between these compartments (67). A novel study mined bulk RNA-seq data from the first trimester decidua and placental villi to identify sexually dimorphic receptor-ligand pairs that could influence cellular interactions in these compartments (69). Subsequent scRNA-seq was utilized to identify the top sexually dimorphic genes within major cell type clusters (e.g., trophoblast, stromal, Hofbauer, APC, and endothelial) as well as placental cell ligands that interact with decidual cell receptors (69). Through this method, 91 sexually dimorphic receptor-ligand pairs were identified as being expressed by cell types at the maternal-fetal interface, which corresponded to processes such as immunomodulation, hormonal regulation, and metabolism (69). Thus, scRNA-seq can also provide evidence of sex-specific differences in cell-cell interactions that are characteristic of healthy pregnancies.
Single-cell landscape of recurrent spontaneous abortion
Comparative studies have applied scRNA-seq technologies to investigate the cellular repertoire and interactions in obstetric disease during early pregnancy, namely recurrent spontaneous abortion (RSA) (71, 73–75, 77). The single-cell investigation of decidual leukocyte subsets indicated an increased presence of T cells in RSA cases (73, 74), which was accompanied by diminished frequencies of specific macrophage (73, 74) and NK subsets (73–75). For example, a CSF1+CD59+ NK cell subset that was prevalent in normal pregnancy was found to be reduced in RSA (75). Moreover, expression and interaction analyses indicated more active pro-inflammatory signaling for decidual leukocytes such as T cells, macrophages, and NK cells in RSA cases compared to controls (73–75). Macrophage interactions with T cells and NK cells are also altered in RSA, as one macrophage subset showed enrichment of processes related to NK cell chemotaxis in healthy pregnancies but shifted towards processes related to T cell chemotaxis in RSA (74). The latter finding was verified using immunofluorescence staining, which revealed more prevalent co-localization of macrophages with NK cells in healthy pregnancy and more co-localization of macrophages with T cells in RSA (74). Thus, altered macrophage interactions with NK cells may result in aberrant promotion of T-cell responses associated with disease in early pregnancy (Figure 1).
In addition to a general state of decidual immune activation, evaluation of the DSC compartment indicated a disease-driven shift in these cells, with several subsets diminishing in RSA patients (71). Notably, a new DSC subset emerged only in RSA cases, which displayed gene expression enriched for cell apoptosis and senescence, potentially indicating abnormal DSC differentiation (71). Consistent with these observations, the developmental trajectories of DSCs were altered in RSA, with one potential differentiation pathway being almost completely absent (71). Moreover, overall decidual cell-cell communications were increased in RSA cases (71). Cellular interaction analyses suggested that DSCs function as a central hub that communicates with other cell types in this compartment (71), and thus defective DSC development or function could trigger a chain of adverse events implicated in RSA. This concept is further supported by the reported reduced expression of the MYC-associated factor X (MAX) in DSCs from RSA patients, which is implicated in proper decidualization and may indicate that impairment of this process is a contributing factor to disease development (77).
The above investigations demonstrate the capacity for single-cell technologies to provide novel insights into pregnancy disease such as RSA, which remains a challenge for obstetrics.
The second trimester
The second trimester represents a period of continued growth and development, characterized by a homeostatic environment at the maternal-fetal interface that is maintained by the cells present in these compartments (124–127). One exploratory study of placental cell types included second trimester samples as a means of comparing changes in single-cell composition throughout pregnancy (66). In this study, EVTs derived from the second-trimester placental villi formed two distinct clusters, one with enrichment of wound and cell adhesion processes and the other with enrichment of growth, response to stimulus, and hormone responses (66). Moreover, such EVTs represented the endpoint of the predicted differentiation pathway when combined with pseudotime trajectory analysis of first-trimester trophoblast subsets (66). These findings confirms that trophoblast differentiation continues throughout the second trimester. More recently, the developmental trajectories of CTBs were specifically explored in the human second trimester placenta to avoid the inflammation and apoptosis associated with parturition in late pregnancy (128). In this study, both the smooth chorion (i.e., the chorion layer of the chorioamniotic membranes) and the villous chorion (within the placenta) were compared using scRNA-seq (128). Consistent with other reports, major cell types included CTB, EVT, immune cells, stromal cells, and a small subset of epithelial cells (128). Notably, a subset of CTBs was identified as being unique to the smooth chorion, termed SC-CTBs, and showed high expression of cytokeratins required for tissue integrity, as verified by immunofluorescence imaging (128). Trajectory analyses revealed that SC-CTBs in the smooth chorion and SCTs in the villous chorion shared a common progenitor (128). Notably, smooth chorion CTBs were found to exhibit greater inhibitory effects on EVTs compared to villous chorion CTBs in an ex vivo setting, and this affect was attributed to the SC-CTBs present in this compartment (128). Taken together, these findings point to distinct trophoblast cell types present in each compartment of the maternal-fetal interface that exhibit differential functionality despite originating from a shared progenitor.
Another recent investigation of the human maternal-fetal interface during the second trimester performed mass cytometry (CyTOF) to survey the leukocyte population present in this compartment (84). This technique was coupled with IMC and immunofluorescence staining to demonstrate the spatial distribution of specific cell types within the placental tissue as well as validate tissue residence by localizing such cells outside of the fetal vasculature (84). CD4+ T cells and NK cells were observed to be more abundant in the decidua, with macrophages being more highly present in the placental villi (84). Specifically, the placental villi were enriched for subsets of CCR7− macrophages and DCs as well as CD8+CD69− T cells, suggesting a relatively inactive status of these cells (84). Deeper evaluation of expressed markers indicated subsets of CD163hi and CD163lo macrophages in the placental villi, with the former likely to be a classical Hofbauer subset, and evaluation of chemokine receptors on placental cells showed reduced expression of several of these markers, which was confirmed by the bulk RNA-seq data (84). The majority of placental T cells were found to be CD8+ memory T cells, with marker expression suggestive of Th2 differentiation, while CD4+ Tregs were more abundant in the decidua (84). A later IMC-based study demonstrated similar findings and further indicated that the immune cell compartment undergoes changes in mid-pregnancy, such as a gradual increase in myeloid cells coupled with declining NK cell abundance (113). As noted above, such a decline in decidual NK cells included a substantial decrease in the abundance of the CD69+ dNK1 subset (113). Similarly, specific myeloid subsets showed distinct trends throughout gestation, supporting the concept that the overall changes in total NK cell or myeloid populations may not reflect the changes in individual subsets (113). Together, these observations provide an overview of the diverse immune cell landscape in the placental villi, and provide further evidence of a largely homeostatic microenvironment at the maternal-fetal interface during mid-gestation.
Several investigations have also explored the murine placental tissues during mid-gestation to unravel the cellular differentiation taking place at this time (81–83, 85). One study performed single-nuclei RNA-seq (snRNA-seq) to ensure adequate representation of SCTs, and evaluated the murine placenta at days 9.5, 10.5, 12.5, and 14.5 of gestation (82). In this study, which was focused on placental labyrinth development, the trophoblast population was dissected to identify sinusoidal trophoblast giant cells, SCT subsets including precursors, glycogen cells, spongiotrophoblasts and their precursors, and junctional zone precursors (82). Such findings expand on an earlier report that utilized scRNA-seq of the day 9.5 placenta to identify major clusters of progenitor trophoblasts and trophoblast giant cells in addition to DSCs, NK cells, and endothelial cells (83). Pseudotime trajectory analysis was implemented to describe several distinct differentiation pathways that included precursor, intermediate, and endpoint states as well as the key genes participating in each pathway (82). Moreover, based on the multiple sampled time points, additional temporal input was used to generate a map of placental development during mid-gestation and infer the functional roles of each identified cell type in this process (82). In a separate investigation of the day 10.5 placenta, trajectory analysis were extended to also include Hofbauer cell subsets, demonstrating a clear progression of Monocyte > Hofbauer cluster 1 > Hofbauer cluster 2 (85). Consistent with the homeostatic microenvironment in early (32) and late (37) gestation, the most abundant placental cell-cell interactions at day 10.5 were between endothelial cell, pericyte, decidual cell, and labyrinth SCT pairs (85). Finally, another investigation focused on the fetal cells present in the placenta in mid-gestation by using a mating strategy wherein transgenic GFP+ male mice were mated with wild type females (81). This study showed that hematopoietic precursors exist in the placenta that can give rise to a subset of fetal macrophages (81), shedding new light on cellular ontogeny in this organ.
Collectively, these studies elucidate the placental developmental processes taking place during mid-pregnancy, and highlight this homeostatic period as an area for future investigations.
The third trimester and delivery
Placental cellular immunobiology
Single-cell technologies have allowed for an unprecedented level of insight into the cellular populations and networks at the maternal-fetal interface in the third trimester, both in the presence and absence of labor. Indeed, two pioneering studies focused on the cellular dynamics of the human placenta derived from term cesarean section deliveries to provide a single-cell atlas of this organ (78, 79). The first study focused on placental trophoblast cell types as well as a small subset of maternal immune cells, and incorporated these data together with sequencing of SCTs collected by laser microdissection, primary undifferentiated endometrial stromal fibroblast cells, and primary decidual cells to obtain a combined dataset that is representative of placental cellular composition (78). The trophoblast clusters were assigned identities as VCT subsets and EVTs, with EVTs showing gene expression signatures associated with modulation of extracellular matrix, vascularization, and immune pathways (78). Interestingly, it was noted that SCTs displayed low-level expression of MHC-II, which is in contrast to other trophoblast subsets (78). The small maternal immune cell cluster was identified as DCs, which could be contaminating cells derived from the uterus (78). In a subsequent study, placental cell type clusters were identified that corresponded to populations of vascular endothelial cells, vascular smooth muscle cells, villous stromal cells, macrophages, trophoblasts, DSCs, DCs, T cells, and an erythrocytic subset, with the trophoblast cluster being further divided into EVTs, CTBs, and SCTs (79). By including paired biopsies sampled proximally and distally to the umbilical cord insertion point, it was shown that there is spatial heterogeneity that is reflected in the relative abundance of cell types such as DSCs and endothelial cells (79), which may be due to the convergence of chorionic arteries and veins at the umbilical cord (129). Analysis of the placental cell-cell communications network indicated likely interactions between adjacent maternal and placental cell types, with DSCs showing a high amount of signaling to and from SCT and EVT (78). Inter-trophoblast communication was also highlighted, as each CTB subset displayed putative interactions with SCT and EVT (78). Notably, by contrasting cell-cell communications using the undifferentiated endometrial stromal fibroblast cells and DSCs, it was found that decidualization enhances signaling potential between DSCs and the fetal trophoblast (78). Together, these two pioneer studies provided important new insights into placental cellular interactions as well as valuable single-cell datasets that have been leveraged by later investigations (80, 130–132).
To enhance the translational value of single-cell placental dynamics, Tsang et al. extrapolated the cellular signatures to the maternal circulating cell-free RNA (79). This novel method was based on studies showing that cell-free DNA and RNA derived from the fetus/placenta are found in the maternal circulation (133–140). Using this approach, it was found that expression profiles corresponding to DSCs, endothelial cells, smooth muscle cells, stromal cells, and monocytes steadily increased in the maternal circulation throughout pregnancy (79). By contrast, EVT, SCT, and B-cell profiles tended to decrease, particularly towards the end of gestation (79). Finally, the overall T-cell population signature decreased in mid-pregnancy and then increased in the third trimester and post-partum period (79), which is in line with the concept that T-cell activation is implicated in the inflammatory milieu that accompanies term parturition at the maternal-fetal interface (38–40, 43, 45, 50, 141).
A recent report focused on in vitro-expanded placental mesenchymal stem/stromal cells demonstrated multiple subsets contained within this population, including some with immunomodulatory gene expression signatures (142). Moreover, these cells expressed cytokines such as CCL2, immunomodulatory factors such as IFITMs, and the regulatory factor PRDM1 (BLIMP-1), and scATAC-seq indicated high chromatin accessibility of immune regions (142), further supporting the involvement of placental mesenchymal stem/stromal cells in immune processes. Moreover, the findings generated in this study support the potential application of placental mesenchymal stem/stromal cells for regenerative or immunomodulatory cell-based therapies.
To date, only one report has provided comparative scRNA-seq analysis of the human placental villi in the presence and absence of spontaneous labor at term (80). In particular, relative differences in the proportions of CTB subsets, activated T cells, monocytes, and macrophages are observed between the labor and non-labor placental villi, with such immune subsets including cells of maternal and fetal origin (80). Moreover, term labor was associated with substantial changes in gene expression across multiple placental cell types including macrophages, monocytes, stromal cells, EVTs, and CTB (80). Such expression changes were enriched for labor-associated terms, such as vascular smooth muscle contraction in the fibroblast subset and cell cycle/metabolism in EVTs (80). Moreover, by applying an approach similar to that reported in (79), gene signatures corresponding to placental cell types could be monitored in the maternal circulation throughout pregnancy, and signatures of NK cells and T cells were found to be enhanced with term labor (80). Thus, such analyses not only provide insight into the labor-specific placental changes that occur at the single-cell level, but provide a potential means of monitoring pregnancy and labor in the maternal circulation.
Decidual cellular immunology
The cellular composition of the decidua undergoes modification throughout gestation in preparation for the inflammatory process of labor (45, 47, 141, 143). Single-cell surveys of the decidua obtained from term cesarean deliveries characterized the overall proportions of cell types in this compartment, including a substantial fraction of T cells (both resting and activated), NK cells, DSCs, endothelial cells, and fibroblasts as well as invasive EVTs, and subsets such as macrophages showed substantial labor-associated changes in gene expression (80, 87) (Figure 2). These data are in line with prior cytological surveys of the decidua showing that leukocytes are attracted to this compartment prior to the onset of labor (40, 43, 49, 141, 144–147). Moreover, a new subset of lymphatic endothelial decidual (LED) cell was described in the chorioamniotic membranes that displayed an expression profile enriched for cellular interactions and adhesion (80). This observation suggested that LEDs present in the chorioamniotic membranes may be functionally mediating the influx of immune cells into this compartment during the process of labor (80). Indeed, immunofluorescence staining of the chorioamniotic membranes revealed the co-expression of the lymphatic marker LYVE1 and the endothelial marker CD31, demonstrating the presence of lymphatic vessels in the decidua parietalis (80). Consistently, a single-cell study focused on decidual endothelial subsets in term non-labor deliveries identified five distinct cell clusters with differing expression profiles, two of which were enriched for cell adhesion processes (86). Together, these studies suggested that decidual endothelial cells, including the novel LED cell type, can contribute to the accumulation of infiltrating lymphocytes into the maternal-fetal interface in preparation for and during term parturition.
Figure 2. The single-cell immunobiology of the maternal-fetal interface in late pregnancy, preterm parturition, and preeclampsia.
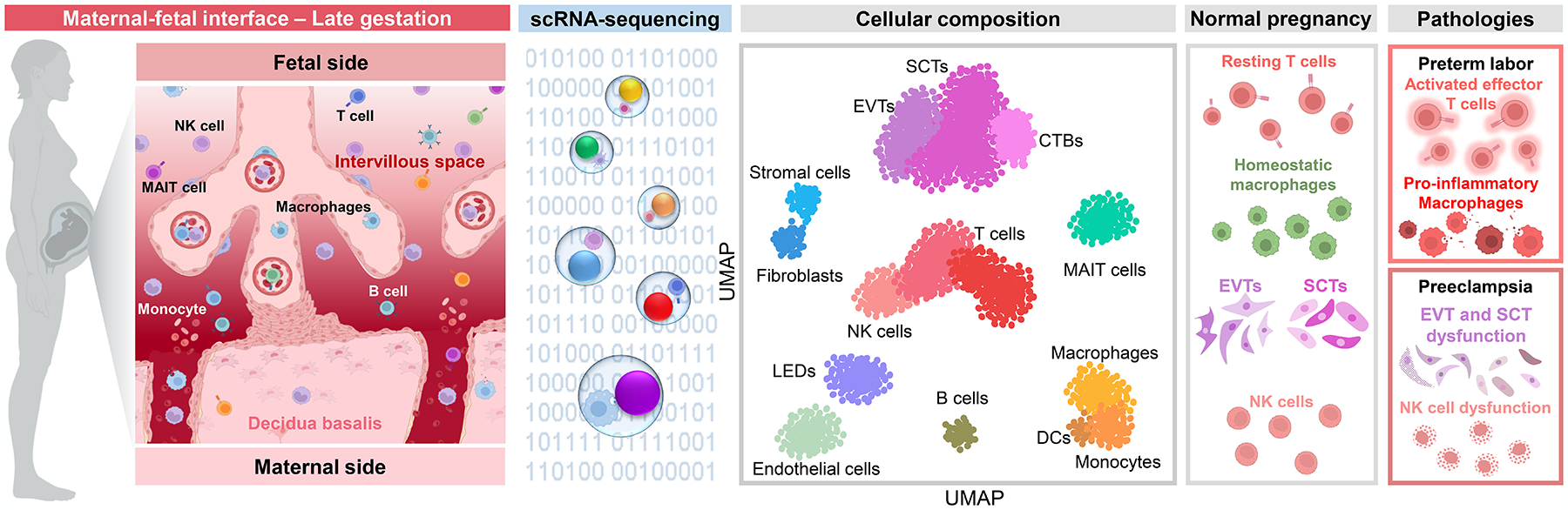
As the end of pregnancy nears, the maternal-fetal interface undergoes changes in cellular composition and gene expression profiles in preparation of labor onset. The decidual lymphocyte population is predominantly composed of T cells, which include a subset of mucosal-associated invariant T cells (as visualized in the UMAP plot based on data reported by Pique-Regi et al. 2019). Decidual stromal and endothelial cells, including the recently described lymphoid endothelial decidual cells, undergo shifts in their expression profiles to display cellular processes and interaction networks that support leukocyte infiltration and activation in the maternal-fetal interface to promote labor. Placental trophoblast cell types also display labor-specific alterations in their signaling profiles, and cell-cell communication between placental and decidual cell types is shown to be enhanced with labor, indicative of an overall increase in intercellular signaling across tissue compartments at the maternal-fetal interface. While preterm labor also involves altered trophoblast signaling, many impacted genes are regulated in the opposite direction compared to normal labor at term, suggesting the distinct activation of placental cell types. Importantly, preterm labor is characterized by the pathological aberrant activation of effector T cells and monocytes/macrophages at the maternal-fetal interface. In preeclampsia, placental trophoblast and fibroblast cell types show enrichment of genes associated with inflammation, oxidative stress, and angiogenesis, indicative of dysregulated signaling that could contribute to placental dysfunction. Such single-cell transcriptomic changes are much more apparent in placentas from cases of early-onset preeclampsia, which is consistent with this disease subset being largely placenta-driven. A number of reports have also implicated dysfunction of decidual NK cells in the pathogenesis of preeclampsia, which could result from functional variations driven by maternal killer cell immunoglobulin-like receptor (KIR) and fetal HLA-C interactions. Abbreviations used: SCT, syncytiotrophoblast; CTB, cytotrophoblast; EVT, extravillous trophoblast; MAIT cell, mucosal-associated invariant T cell, NK cell, natural killer cell; DC, dendritic cell; LED, lymphoid endothelial decidual cell; UMAP, uniform manifold approximation and projection. Figure created using BioRender.
Mucosal-associated invariant T (MAIT) cells, an innate-like T-cell subset expressing a restricted TCR (148), have also been detected at the maternal-fetal interface (149–151); yet, the expression profiles and functionality of these cells has been underexplored. A targeted investigation utilized combined flow cytometry, CITE-seq, and scRNA-seq approaches to characterize MAIT cells in the decidua basalis and parietalis, demonstrating that the majority of these cells displayed an effector memory phenotype (152). Decidual MAIT cells exhibited gene expression profiles distinct from those found in the periphery, including genes involved in immune suppression and cell migration (152). Key differentiation factors upregulated in MAIT cells compared to conventional T cells included PLZF and EOMES, and in vitro stimulation resulted in secretion of IFNγ and TNF together with granzyme B, indicating that these cells are functional and can participate in maternal-fetal immune responses (152).
Recent investigations have also compared the changes in cellular composition and cell-cell signaling between decidual tissues derived from labor and non-labor deliveries to evaluate the participation of individual subsets in the inflammatory process of parturition. Spontaneous term labor was shown to result in the increased prevalence of activated T cells, monocytes, macrophages, and DSCs in the decidua basalis compared to term deliveries without labor (80). Moreover, IMC analysis of the maternal-fetal interface at term indicated increased abundance of myeloid cells and T cells compared to second trimester samples (113). Such changes are consistent with prior studies indicating an influx of immune cells to the maternal-fetal interface during parturition (40, 43, 47, 141, 144, 146).
More prominent than changes in cell type proportions are the transcriptomic changes that occur to facilitate labor. Macrophages, stromal cells, monocytes, T cells, and EVTs, among others, showed drastic differential gene expression during term labor compared to their non-labor counterparts, and such genes were enriched for inflammatory and immunomodulatory pathways (80). Similarly, decidual endothelial cell clusters each showed labor-specific enrichment of inflammatory pathways such as IL-17 signaling, leukocyte differentiation, myeloid differentiation, and cytokine-related terms (86), as did DSC and decidual EVT subsets (87), suggesting that decidual immune and tissue cell subsets propagate inflammation to drive parturition. Such changes in the transcriptomic profiles of decidual cells manifest in altered cell-cell communications, as indicated by numerous enhanced signaling pathways such as IL-1β – IL-1R1, IL-6 – IL-6R, TNFSF14 – LTBR, and multiple chemokine – chemokine receptor pathways (88). Rather than solely regulating new pathways, labor also seems to involve the continued upregulation or downregulation of already-modulated signaling pathways (88), suggesting that transcriptomic changes implicated in labor may be initiated earlier in gestation. In particular, T cells undergo significant shifts in intercellular communications with other decidual cells, involving pathways such as Notch, NF-κB, MAPK, Jak-STAT, and chemokine signaling, among others (88). However, these underlying labor-associated transcriptomic changes did not seem to impact the TCR clonality and diversity within the decidua (88), consistent with the concept that the decidual T-cell compartment is primarily composed of memory T cells (43, 84, 152). Together, these data point to a decidual inflammatory response implicated in labor that involves transcriptomic changes in specific cell types from the innate and adaptive limbs of the immune system.
The use of scRNA-seq to decipher the mechanisms leading to preterm labor
Spontaneous preterm labor is not simply the premature activation of the common pathway of parturition, but rather represents a pathological process that is distinct from normal labor at term (153). Unravelling the molecular mechanisms underlying spontaneous preterm labor is therefore an ongoing endeavor within our group. Thus, we have leveraged scRNA-seq to compare and contrast the cellular composition and transcriptomic profiles of the decidua, placenta, and chorioamniotic membranes in term and preterm labor to improve the understanding and potential prediction of this obstetrical syndrome (80) (Figure 2). Compared to term labor, preterm labor involves substantial changes in gene expression within the EVT and CTB populations, potentially indicating a distinct response in these cell types (80). Moreover, within preterm labor-affected subsets such as EVTs and CTBs, specific genes were identified that showed opposite direction of change compared to term labor, indicating that distinct cellular mechanisms are involved in these processes (80). To extend the clinical relevance of our findings, we applied our scRNA-seq-derived signatures to the cellular transcriptome of the maternal circulation and showed that signatures of maternal macrophages, monocytes, and activated T cells were elevated in women who underwent spontaneous preterm labor compared to gestational age-matched controls, supporting a role for intravascular immune activation in the pathophysiology of preterm labor (80). Thus, the maternal circulation may provide a useful window that can be used to monitor preterm labor-specific events at the maternal-fetal interface.
More recently, we have undertaken the investigation of the cellular interactions implicated in pathological labor using a murine model of preterm birth induced by the intra-amniotic inoculation of E. coli (90). We evaluated the key tissues implicated in the common pathway of labor (i.e., the uterus, decidua, and cervix) at single-cell resolution and demonstrated that preterm labor affects the cellularity of the uterus, decidua, and cervix through immune cell infiltration and altered transcriptomic profiles of non-immune cell types in a tissue-specific manner (90). In the uterus, both innate and adaptive immune cell subsets (e.g., neutrophils, macrophages, DCs, NK cells, and T cells) contributed to pathways implicated in preterm labor, such as cytokine and chemokine signaling, as did non-immune subsets (e.g., fibroblasts, stromal cells, epithelial cells, smooth muscle cells, and endothelial cells), and the interaction strength among cell types was altered with preterm labor (90). Similar changes in cell-cell signaling were observed in the decidua; yet, unique pathways such as IL-17 were also revealed in this tissue (90). The cellularity of the cervix predominantly involved epithelial cells, which showed increased incoming interactions from other tissue cell subsets during preterm labor (90). Importantly, although specific cell types displayed consistent signaling across tissues, each also exhibited tissue-specific processes with preterm labor, indicating unique functions tailored to the tissue microenvironment (90). Together, these findings provide novel insight into the cellular changes taking place in the murine reproductive tissues during preterm labor and lead to premature delivery.
Placental scRNA-seq sheds light into the pathophysiology of preeclampsia
Preeclampsia is primarily a placental and intravascular disease (154), and thus this tissue has been the target of single-cell investigations aimed at uncovering the molecular mechanisms underlying the development of this obstetrical syndrome (79, 155–157). One of the first comprehensive scRNA-seq studies integrated placenta-derived single-cell signatures with bulk transcriptomic data derived from the plasma cell-free RNA of women with early-onset preeclampsia and healthy pregnancies (79). Signatures corresponding to decidual cells, endothelial cells, and EVTs were upregulated in the maternal plasma with preeclampsia, whereas the SCT signature was reduced (79). Analysis of the transcriptional heterogeneity of genes involved in cell migration, cell proliferation and apoptosis were more variable in preeclamptic patients compared to normal term deliveries; moreover, genes annotated to cell death had overall higher expression in preeclampsia (79), which is consistent with prior associations between this disease and trophoblast apoptosis (158–162) (Figure 2). Consistently, the direct scRNA-seq comparison of preeclamptic and healthy placentas indicated disease-driven enrichment of genes annotated to oxidative stress and inflammation in EVTs (155, 157), and those annotated to cell cycle and protein folding in SCTs (157). Moreover, clustering of differentially regulated genes revealed a module with reduced activity in preeclampsia that was related to cytokines, regulation of cell death, and differentiation (157). Two transcription factors, CEBPB and GTF2B, were identified as being greatly reduced in the disease state, and thus could be implicated in the trophoblast dysfunction characteristic of preeclampsia (157).
Preeclampsia has been traditionally classified into two subsets, with each being characterized by distinct pathophysiology: early-onset and late-onset (154, 163–165). To unravel cellular changes that may distinguish early- and late-onset preeclampsia, an in silico investigation utilized placental single-cell signatures derived from the study of Vento-Tormo et al. (68) to deconvolute bulk transcriptomic data of placentas from women with either form of the disease (156). The number of genes differentially regulated in early-onset preeclampsia compared to its control group was substantially greater than in late-onset (156), which is consistent with early-onset being largely a placental disease (154, 166–168). Moreover, the direct comparison of early- and late-onset preeclampsia also resulted in a large set of differentially expressed genes, further indicating distinct underlying pathophysiology (156). Deeper investigation of changes at single-cell resolution indicated that EVTs and fibroblasts play a key role in early-onset preeclampsia, displaying dysregulated signaling associated with angiogenesis and fibrosis (156). Importantly, novel trophoblast-derived markers were identified as being upregulated in early-onset preeclampsia, such as Epstein-Barr virus induced 3 (EBI3), and such proteins could be detected as being elevated in the maternal circulation (156). Therefore, the combination of placental single-cell transcriptomics with soluble biomarker discovery can represent a viable approach for the prediction and diagnosis of preeclampsia.
Myometrial cell types and their contributions to spontaneous term labor
The uterine myometrium and the cervix are both key organs in the common pathway of parturition (169–171). Thus, investigation of the cellular processes taking place in these tissues prior to the initiation of labor can provide a baseline measurement that is useful for the study of physiological or pathological labor. A recent study obtained myometrial and cervical biopsies during planned cesarean hysterectomy without evidence of labor in the third trimester and performed scRNA-seq (172). The resulting data were cross-referenced with the Human Protein Atlas to identify major cell type clusters, which included endothelial, epithelial, and stromal cells, smooth muscle cells, and leukocytes (172). Expression of CD74 (HLA class II histocompatibility antigen gamma chain) could distinguish the epithelial, endothelial, and leukocyte clusters from the stromal and smooth muscle cells (172). As CD74 regulates the function of MHC class II molecules (173) and is thought to be mainly restricted to APCs under homeostatic conditions, a potential underlying state of tissue inflammation was proposed that could drive CD74 expression in epithelial/endothelial cells (174, 175). This finding was consistent with the observed upregulation of genes associated with inflammation (172); yet, given that biopsies were taken prior to active labor, such inflammation could represent preparation for the onset of parturition.
We recently undertook a single-cell survey of the human uterine tissue (primarily myometrium) to further investigate the molecular mechanisms underlying the transition from a quiescent to a contractile state prior to term labor (89). Myometrial biopsies were obtained from term deliveries either with or without spontaneous labor for scRNA-seq, from which a total of 24 immune and non-immune cell types were identified, including multiple subsets of smooth muscle cells and macrophages (89). Specifically, we classified three subsets of smooth muscle cell according to their transcriptomic profiles, which were then validated by protein expression (89). We showed that the first smooth muscle subset displayed an expression profile involving smooth muscle contraction, the second included processes related to neutrophil biology (e.g., neutrophil elastase expression), and the third exhibited increased interferon (IFN)-γ signaling, demonstrating the differing roles of these novel subsets in the process of labor (89). Transcriptomic characterization of macrophage subsets and stromal cell types was also reported (89). Comparison of overall changes in cell abundance indicated a substantial labor-specific increase in the presence of stromal cells, endothelial cells, monocytes, decidual cells, and myofibroblasts, which was accompanied by reductions in macrophage and lymphocyte subsets (89). Consistently, the most dramatic labor-driven changes in gene expression were found in stromal cells, endothelial cells, monocytes, and macrophages, which included an overall enrichment of multiple muscle- and contraction-related processes (89). We also evaluated intercellular communications and identified major signaling pathways implicated in labor such as complement, contraction, IL-1, TGF-β, and THY1 (89). In particular, we show that specific myometrial cell types such as decidual cells, EVTs, myofibroblasts, smooth muscle cells, and stromal cells act as receivers of IL-1 (89), which is a considered a master regulator of human parturition (176–179). By comparing scRNA-seq data with bulk transcriptomics of the human myometrium, we demonstrated a high degree of agreement between these datasets, with single-cell technology providing better coverage of differential gene expression (89). Consistent with prior analyses, gene expression signatures corresponding to scRNA-seq-derived myometrial cell types could be monitored in the maternal circulation, allowing for the evaluation of cellular dynamics throughout pregnancy (89). Importantly, such comparison indicated agreement in the labor-specific enrichment of specific cell type signatures, namely monocytes, thereby providing potential biomarkers that could be indicative of labor progression (89).
In addition to providing insight into the cellular changes and interactions associated with term labor, single-cell technology has also been applied to evaluate resident regulatory T cells (Tregs) present in the placental bed (uterine tissues) biopsied after term cesarean delivery (180). Uterine Tregs (uTregs) expressed a core transcriptomic signature consistent with classical Tregs, which was even more pronounced than peripheral Tregs and included expression of FOXP3, CTLA4, and IL2RA (180). In vitro assays confirmed the suppressive functionality of these cells, which was consistent with the observed gene expression/pathway enrichment indicative of activated, effector uTregs (180). By comparing uTregs with publically available Treg datasets from other tissues, it was shown that such cells displayed transcriptomic overlap with tissue- and tumor-infiltrating Tregs (180). These data provide invaluable insight into uTregs that participate in the modulation of local effector T cells to prevent aberrant immune activation during pregnancy.
Together, the studies outlined above point to the uterine myometrium as an underexplored site of cellular interactions and signaling that are necessary for physiological labor at term. Future single-cell investigations may further explore such pathways in the context of pathological labor leading to preterm birth as well as other pregnancy complications.
Single-cell profile of the maternal-fetal interface in patients with COVID-19
Since the emergence of the SARS-CoV-2 virus in late 2019, investigators around the globe have shifted research efforts to uncovering the molecular mechanisms that dictate maternal infection, disease severity, and risk of vertical transmission during pregnancy. Initially, to further this goal, multiple reports undertook in silico analysis of previously generated single-cell data (66, 68, 78) to evaluate the expression of the canonical SARS-CoV-2 cell entry mediators angiotensin converting enzyme 2 (ACE2) (181–183) and transmembrane protease serine 2 (TMPRSS2) (183) at the maternal-fetal interface (92, 94–96, 99, 184, 185). Each of these studies observed independent expression of ACE2 and TMPRSS2 in decidual cells such as DSCs and perivascular cells (92, 96, 184) and/or in placental trophoblast subsets (92, 95, 96, 185). However, viral cell entry requires the co-expression of both ACE2 and TMPRSS2 within the same cell (183), and the evidence for co-expression of such cell entry mediators by placental cells was unclear, given that some studies indicated detectable co-expression (95, 185) while others suggested this was rare (94, 96, 99). Therefore, to provide further clarity in this regard, we undertook a combined approach that utilized: 1) previous single-cell data of the first-trimester maternal-fetal interface (Vento-Tormo et al. 2018 (68)); 2) new single-cell data of the decidua and placenta from an indicated second-trimester hysterectomy; and 3) our previous single-cell dataset of the third-trimester decidua basalis, placental villi, and chorioamniotic membranes (Pique-Regi et al. 2019 (80)) (91). We found that a minimal number of cells co-express ACE2 and TMPRSS2 at the maternal-fetal interface throughout gestation, even with an extremely permissive expression threshold (91). To overcome the limitation of low SCT representation in our single-cell dataset (due to their multinucleated morphology), snRNA-seq of placental tissues was performed and confirmed that co-expression of ACE2 and TMPRSS2 is minimal among these cells (91). Thus, our data support the absence of meaningful co-expression of classical cell entry mediators for SARS-CoV-2 at the maternal-fetal interface throughout pregnancy, a concept that has since been further confirmed (186).
Regardless of the expression patterns of viral cell entry mediators, a central question amidst the COVID-19 pandemic has been whether maternal infection results in a fetal/placental immune response that can lead to adverse pregnancy outcomes and, more importantly, whether the fetus itself is impacted. A study utilized bulk and single-cell transcriptomic analyses of the placental tissues from women diagnosed with COVID-19 to demonstrate upregulation of genes associated with immune response compared to uninfected controls (93). At single-cell resolution, such COVID-19-driven changes included enrichment of cytotoxic molecules in NK cells and signs of T-cell activation, and endothelial cells similarly displayed signs of activation and immune response (93). This finding is consistent with a case report in which scRNA-seq was performed using placental tissues from a pregnant COVID-19 patient who was delivered at 28 weeks of gestation, showing that CD8+ T cells were activated (98). Moreover, cell-cell communication analysis indicated increased interactions between immune cells at the maternal-fetal interface in COVID-19 (93, 98), including between T cells and monocytes/NK cells (93). In light of this evidence pointing to a maternal immune response induced by COVID-19 during pregnancy, we undertook a comprehensive multi-disciplinary investigation of maternal-fetal immunity in SARS-CoV-2-infected pregnant women, most of whom were asymptomatic, and showed using scRNA-seq that maternal T cells and macrophages in the chorioamniotic membranes display substantial changes in gene expression compared to cells from uninfected pregnant women (97). We then compared our T-cell signatures with previously generated single-cell signatures of peripheral T cells from hospitalized COVID-19 patients, and observed a positive correlation between these datasets (97), suggesting that T cells derived from the maternal-fetal interface of pregnant COVID-19 patients display similar characteristics to those found in the circulation of non-pregnant patients with severe disease. Shared genes were enriched for protein translation processes; yet, some differentially expressed genes were unique to maternal-fetal interface-derived T cells (97). The combined differentially expressed genes from the maternal-fetal interface of women with COVID-19 showed enrichment of multiple interferon signaling pathways, indicating that SARS-CoV-2 infection drives an anti-viral immune response even when the virus itself is not present in this compartment (97). Overall, these novel findings support the value of single-cell datasets that can be leveraged to investigate relevant cellular and molecular targets for diseases such as COVID-19, which in turn can inform translational research directed at therapeutic interventions.
Limitations of single-cell technology
The emergence and popularization of single-cell techniques has opened new avenues of scientific discovery across multiple disciplines. Yet, the widespread use of such methods has also highlighted their limitations in comparison to other cellular and molecular tools, such as the inherent discovery-based nature of current single-cell approaches. Moreover, the analysis of scRNA-seq datasets requires advanced computational approaches that rely on algorithmic clustering and associations to determine cell identities and profiles, which can differ based on the platforms and analysis pipelines utilized (as has been extensively reviewed in (187–190) and elsewhere). For this reason, researchers utilizing single-cell approaches have often chosen to validate key findings (e.g., identification of novel cell subsets) by using alternative approaches such as flow cytometry, imaging methods, or animal models. Indeed, we have utilized immunofluorescence imaging to confirm our identification of a potentially novel cell type, lymphoid endothelial decidual (LED) cells, in the human chorioamniotic membranes (80). Similarly, we used the same approach to validate our identification of distinct subsets of smooth muscle cell in the human myometrium based on expression of oxytocin receptor, elastase, or IFNγ (89). For studies in which the identification of novel cell types represents a primary outcome, we consider that additional functional and phenotypic analyses are essential to ensure that such populations do not represent artifacts resulting from data processing and analysis. Overall, we consider that the application of validation techniques is essential for providing confirmatory analysis of discoveries made using exploratory single-cell approaches.
An important consideration for scRNA-seq and other single-cell technologies is the analytical approach that will be utilized to dissect the generated data. A complete discussion of single-cell analysis pipelines and tools is outside of the scope and expertise of this review, and such topics have been covered more extensively elsewhere (187, 188, 190). Yet, we have successfully applied several of the tools described in this review, such as CellChat cell-cell communication analysis (119) and Slingshot trajectory analysis (191), to our single-cell investigations of the human maternal-fetal interface (Figure 3). In particular, an important consideration when studying the placental and decidual tissues at single-cell resolution is the maternal or fetal origin of each cell. To overcome this potential limitation and aid in the interpretation of our scRNA-seq data, we have successfully incorporated maternal and fetal genotyping data into our analysis pipeline (80, 89, 97, 192). Other limitations of scRNA-seq in the context of maternal-fetal immunology include the difficulty of obtaining biopsies for research, for which we have proposed and utilized two different translational solutions: first, we have shown that specific cellular signaling pathways implicated in labor are shared between the human and murine myometrium (90). While validation remains necessary when comparing humans and mice, such findings can lay the groundwork for future single-cell investigations of pregnancy. In parallel, the evaluation of placenta-derived single-cell signatures in the maternal circulation has been shown to represent a viable approach for monitoring pregnancy and its complications in a non-invasive manner, as has been described in this review (79, 80, 89, 131). Taken together, these findings indicate that scRNA-seq represents a translationally-relevant approach for investigation of the maternal-fetal interface; yet, we consider that the careful application of this technology and its analysis is required to ensure the generation of useful and reproducible results.
Figure 3. Overview of applied single-cell analytics tools.
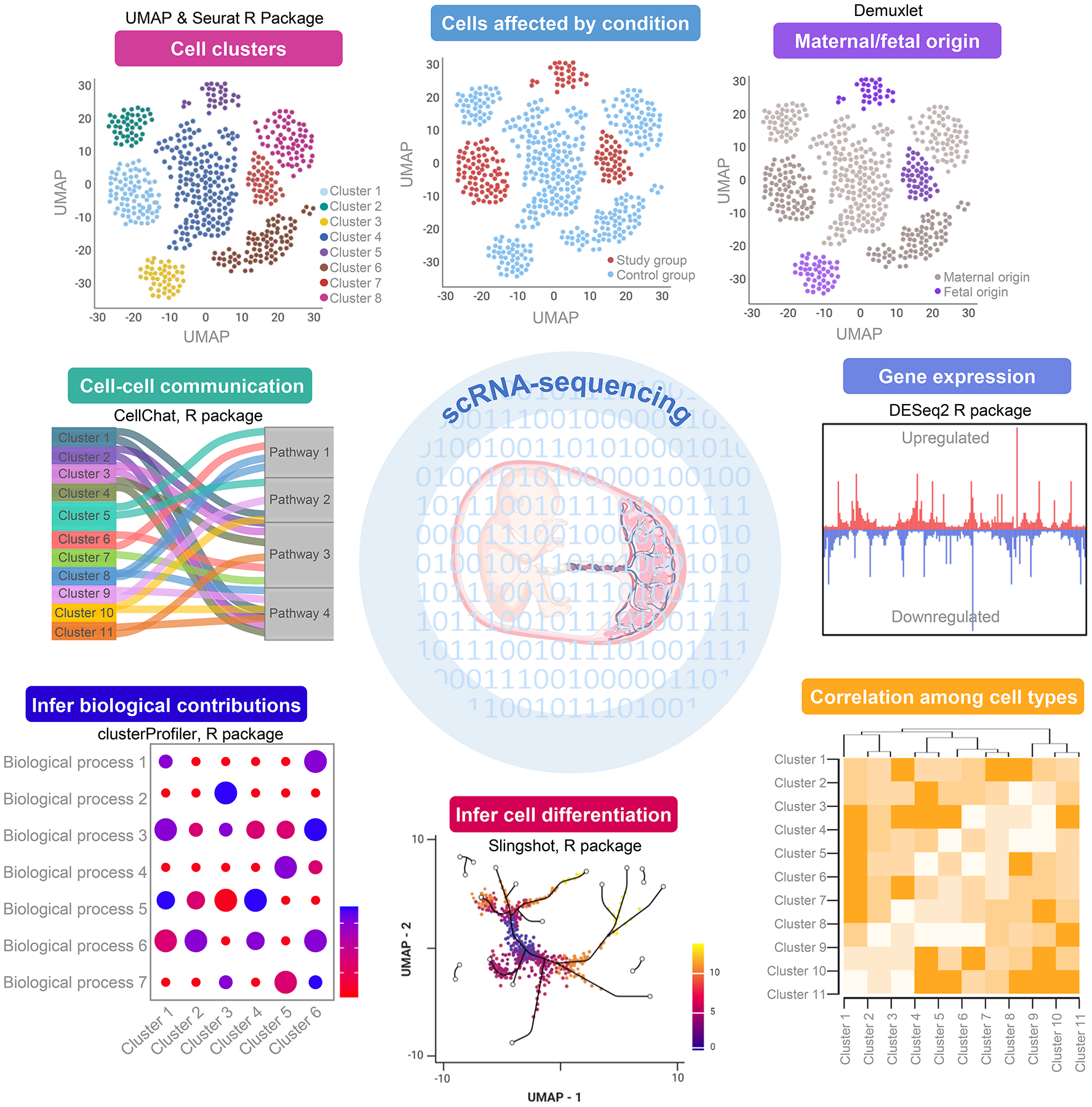
Diagram providing representative images of single-cell data visualization and analysis together with the methods used for their creation. Clockwise direction starting from the upper left pane: Seurat (193, 194) is used to normalize and combine cell count data matrices for subsequent visualization using uniform manifold approximation and projection (UMAP) plots. The demultiplexing of barcoded single-cell libraries allows further visualization according to sample metadata, such as control and study groups. We have further expanded this concept specifically for studies of the maternal-fetal interface to incorporate maternal and fetal genotype data using demuxlet (195), thereby allowing for the identification of maternal and fetal cells as outlined in (Pique-Regi et al. (89)) and (Garcia-Flores et al. (192)). The differential expression of specific genes can be evaluated using DESeq2 (196), which allows for additional downstream analyses such as correlation among cell types, visualization of cell differentiation trajectories using Slingshot (191), or inference of cell type contributions to biological processes using clusterProfiler (197). We have also implemented the recently-developed CellChat (119) to unravel cell-cell communication networks based on the expression of ligands, receptor subunits, cofactors, and other related molecules by each cell type. These tools and visual representations can represent a useful starting point for researchers looking to apply single-cell technology to their investigation of the maternal-fetal interface. Figure created using BioRender.
Conclusions
Collectively, the reports summarized here emphasize the value of single-cell technology for characterizing the cellular dynamics and interactions at the maternal-fetal interface. Given the cellular heterogeneity of the decidua, placenta, and reproductive tissues, “omics” techniques that provide information at single-cell resolution are rapidly becoming the new standard for investigations of the molecular mechanisms underlying developmental processes, physiological and pathological labor, and other obstetrical diseases. The findings reviewed herein provide a rich framework of single-cell data, generated at different gestational time points and under various conditions, which can be a starting point for future studies. Importantly, we recognize that there are often disparities between studies that can result from differences in technologies, individual platforms, and cell-type classification methods, among other variables. Thus, given the abundance of single-cell datasets that have been generated from the maternal-fetal unit, there is potential for unified analyses that integrate such data under consistent cell-type annotations and analysis pipelines to represent single-cell dynamics at different time points throughout pregnancy in a comparable manner. In summary, this review demonstrates the power of single-cell technologies to uncover new cellular immunobiological pathways and interactions that may have translational relevance for pregnancy disease, and it is our hope that this summary can act as a catalyst for future investigations of the maternal-fetal interface.
Funding
This research was supported by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS) under Contract No. HHSN275201300006C. This research was also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. R.R. has contributed to this work as part of his official duties as an employee of the United States Federal Government. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Abbreviations
- APC
antigen-presenting cell
- ATAC-seq
assay for transposase-accessible chromatin with high-throughput sequencing
- CITE-seq
cellular indexing of transcriptomes and epitopes by sequencing
- CTB
cytotrophoblast
- CyTOF
cytometry by time-of-flight
- DC
dendritic cell
- DSC
decidual stromal cell
- EVT
extravillous trophoblast
- HLA
human leukocyte antigen
- IMC
imaging mass cytometry
- IL
interleukin
- ILC
innate lymphoid cell
- KIR
killer immunoglobulin-like receptor
- LED
lymphoid endothelial decidual cell
- MAIT cell
mucosal-associated invariant T cell
- MSC
mesenchymal stromal cell
- MHC
major histocompatibility complex
- NK cell
natural killer cell
- RSA
recurrent spontaneous abortion
- scRNA-seq
single-cell RNA-sequencing
- SCT
syncytiotrophoblast (termed “STB” in some studies)
- snRNA-seq
single-nucleus RNA-sequencing
- TCR
T-cell receptor
- Treg
regulatory T cell
- UMAP
uniform manifold approximation and projection
- VCT
villous cytotrophoblast
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Cunningham FG, Leveno KJ, Dashe JS, Hoffman BL, Spong CY, and Casey BM. 2022. Embryogenesis and Fetal Development. In Williams Obstetrics, 26e. McGraw Hill, New York, NY. [Google Scholar]
- 2.Norwitz ER, Schust DJ, and Fisher SJ. 2001. Implantation and the survival of early pregnancy. N Engl J Med 345: 1400–1408. [DOI] [PubMed] [Google Scholar]
- 3.Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, and Fisher SJ. 2004. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest 114: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffett A, and Loke C. 2006. Immunology of placentation in eutherian mammals. Nat Rev Immunol 6: 584–594. [DOI] [PubMed] [Google Scholar]
- 5.Cha J, Sun X, and Dey SK. 2012. Mechanisms of implantation: strategies for successful pregnancy. Nat Med 18: 1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arck PC, and Hecher K. 2013. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med 19: 548–556. [DOI] [PubMed] [Google Scholar]
- 7.Lash GE 2015. Molecular Cross-Talk at the Feto-Maternal Interface. Cold Spring Harb Perspect Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith OW, Chavan AR, Protopapas S, Maziarz J, Romero R, and Wagner GP. 2017. Embryo implantation evolved from an ancestral inflammatory attachment reaction. Proc Natl Acad Sci U S A 114: E6566–e6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ander SE, Diamond MS, and Coyne CB. 2019. Immune responses at the maternal-fetal interface. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petroff MG 2005. Immune interactions at the maternal-fetal interface. J Reprod Immunol 68: 1–13. [DOI] [PubMed] [Google Scholar]
- 11.Sargent IL, Borzychowski AM, and Redman CW. 2006. NK cells and human pregnancy--an inflammatory view. Trends Immunol 27: 399–404. [DOI] [PubMed] [Google Scholar]
- 12.Erlebacher A 2013. Immunology of the maternal-fetal interface. Annu Rev Immunol 31: 387–411. [DOI] [PubMed] [Google Scholar]
- 13.Mori M, Bogdan A, Balassa T, Csabai T, and Szekeres-Bartho J. 2016. The decidua-the maternal bed embracing the embryo-maintains the pregnancy. Semin Immunopathol 38: 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tersigni C, Redman CW, Dragovic R, Tannetta D, Scambia G, Di Simone N, Sargent I, and Vatish M. 2018. HLA-DR is aberrantly expressed at feto-maternal interface in pre-eclampsia. J Reprod Immunol 129: 48–52. [DOI] [PubMed] [Google Scholar]
- 15.Holder B, Aplin JD, Gomez-Lopez N, Heazell AEP, James JL, Jones CJP, Jones H, Lewis RM, Mor G, Roberts CT, Robertson SA, and Zenclussen AC. 2021. ‘Fetal side’ of the placenta: anatomical mis-annotation of carbon particle ‘transfer’ across the human placenta. Nat Commun 12: 7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gellersen B, Brosens IA, and Brosens JJ. 2007. Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med 25: 445–453. [DOI] [PubMed] [Google Scholar]
- 17.Shima T, Sasaki Y, Itoh M, Nakashima A, Ishii N, Sugamura K, and Saito S. 2010. Regulatory T cells are necessary for implantation and maintenance of early pregnancy but not late pregnancy in allogeneic mice. J Reprod Immunol 85: 121–129. [DOI] [PubMed] [Google Scholar]
- 18.Chen T, Darrasse-Jèze G, Bergot AS, Courau T, Churlaud G, Valdivia K, Strominger JL, Ruocco MG, Chaouat G, and Klatzmann D. 2013. Self-specific memory regulatory T cells protect embryos at implantation in mice. J Immunol 191: 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shima T, Inada K, Nakashima A, Ushijima A, Ito M, Yoshino O, and Saito S. 2015. Paternal antigen-specific proliferating regulatory T cells are increased in uterine-draining lymph nodes just before implantation and in pregnant uterus just after implantation by seminal plasma-priming in allogeneic mouse pregnancy. J Reprod Immunol 108: 72–82. [DOI] [PubMed] [Google Scholar]
- 20.Robertson SA, Care AS, and Moldenhauer LM. 2018. Regulatory T cells in embryo implantation and the immune response to pregnancy. J Clin Invest 128: 4224–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher A, and Zenclussen AC. 2019. Human Chorionic Gonadotropin-Mediated Immune Responses That Facilitate Embryo Implantation and Placentation. Front Immunol 10: 2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann AP, Gerber SA, and Croy BA. 2014. Uterine natural killer cells pace early development of mouse decidua basalis. Mol Hum Reprod 20: 66–76. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, Zheng Q, and Jin L. 2019. Dynamic Function and Composition Changes of Immune Cells During Normal and Pathological Pregnancy at the Maternal-Fetal Interface. Front Immunol 10: 2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaouat G, Voisin GA, Escalier D, and Robert P. 1979. Facilitation reaction (enhancing antibodies and suppressor cells) and rejection reaction (sensitized cells) from the mother to the paternal antigens of the conceptus. Clin Exp Immunol 35: 13–24. [PMC free article] [PubMed] [Google Scholar]
- 25.Bonney EA, and Onyekwuluje J. 2003. The H-Y response in mid-gestation and long after delivery in mice primed before pregnancy. Immunol Invest 32: 71–81. [DOI] [PubMed] [Google Scholar]
- 26.Aluvihare VR, Kallikourdis M, and Betz AG. 2004. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol 5: 266–271. [DOI] [PubMed] [Google Scholar]
- 27.Zenclussen AC, Gerlof K, Zenclussen ML, Sollwedel A, Bertoja AZ, Ritter T, Kotsch K, Leber J, and Volk HD. 2005. Abnormal T-cell reactivity against paternal antigens in spontaneous abortion: adoptive transfer of pregnancy-induced CD4+CD25+ T regulatory cells prevents fetal rejection in a murine abortion model. Am J Pathol 166: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson SA, Guerin LR, Moldenhauer LM, and Hayball JD. 2009. Activating T regulatory cells for tolerance in early pregnancy - the contribution of seminal fluid. J Reprod Immunol 83: 109–116. [DOI] [PubMed] [Google Scholar]
- 29.Kahn DA, and Baltimore D. 2010. Pregnancy induces a fetal antigen-specific maternal T regulatory cell response that contributes to tolerance. Proc Natl Acad Sci U S A 107: 9299–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowe JH, Ertelt JM, Xin L, and Way SS. 2012. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature 490: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, and Rudensky AY. 2012. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 150: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Care AS, Diener KR, Jasper MJ, Brown HM, Ingman WV, and Robertson SA. 2013. Macrophages regulate corpus luteum development during embryo implantation in mice. J Clin Invest 123: 3472–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang TT, Chaturvedi V, Ertelt JM, Kinder JM, Clark DR, Valent AM, Xin L, and Way SS. 2014. Regulatory T cells: new keys for further unlocking the enigma of fetal tolerance and pregnancy complications. J Immunol 192: 4949–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svensson-Arvelund J, Mehta RB, Lindau R, Mirrasekhian E, Rodriguez-Martinez H, Berg G, Lash GE, Jenmalm MC, and Ernerudh J. 2015. The human fetal placenta promotes tolerance against the semiallogeneic fetus by inducing regulatory T cells and homeostatic M2 macrophages. J Immunol 194: 1534–1544. [DOI] [PubMed] [Google Scholar]
- 35.Bonney EA 2016. Immune Regulation in Pregnancy: A Matter of Perspective? Obstet Gynecol Clin North Am 43: 679–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Lopez N, Arenas-Hernandez M, Romero R, Miller D, Garcia-Flores V, Leng Y, Xu Y, Galaz J, Hassan SS, Hsu CD, Tse H, Sanchez-Torres C, Done B, and Tarca AL. 2020. Regulatory T Cells Play a Role in a Subset of Idiopathic Preterm Labor/Birth and Adverse Neonatal Outcomes. Cell Rep 32: 107874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez-Lopez N, Garcia-Flores V, Chin PY, Groome HM, Bijland MT, Diener KR, Romero R, and Robertson SA. 2021. Macrophages exert homeostatic actions in pregnancy to protect against preterm birth and fetal inflammatory injury. JCI Insight 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sindram-Trujillo AP, Scherjon SA, van Hulst-van Miert PP, Kanhai HH, Roelen DL, and Claas FH. 2004. Comparison of decidual leukocytes following spontaneous vaginal delivery and elective cesarean section in uncomplicated human term pregnancy. J Reprod Immunol 62: 125–137. [DOI] [PubMed] [Google Scholar]
- 39.Osman I, Young A, Jordan F, Greer IA, and Norman JE. 2006. Leukocyte density and proinflammatory mediator expression in regional human fetal membranes and decidua before and during labor at term. J Soc Gynecol Investig 13: 97–103. [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Lopez N, Estrada-Gutierrez G, Jimenez-Zamudio L, Vega-Sanchez R, and Vadillo-Ortega F. 2009. Fetal membranes exhibit selective leukocyte chemotaxic activity during human labor. J Reprod Immunol 80: 122–131. [DOI] [PubMed] [Google Scholar]
- 41.Gomez-Lopez N, Vadillo-Perez L, Hernandez-Carbajal A, Godines-Enriquez M, Olson DM, and Vadillo-Ortega F. 2011. Specific inflammatory microenvironments in the zones of the fetal membranes at term delivery. Am J Obstet Gynecol 205: 235.e215–224. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Lopez N, Vadillo-Perez L, Nessim S, Olson DM, and Vadillo-Ortega F. 2011. Choriodecidua and amnion exhibit selective leukocyte chemotaxis during term human labor. Am J Obstet Gynecol 204: 364.e369–316. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Lopez N, Vega-Sanchez R, Castillo-Castrejon M, Romero R, Cubeiro-Arreola K, and Vadillo-Ortega F. 2013. Evidence for a role for the adaptive immune response in human term parturition. Am J Reprod Immunol 69: 212–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leng Y, Romero R, Xu Y, Galaz J, Slutsky R, Arenas-Hernandez M, Garcia-Flores V, Motomura K, Hassan SS, Reboldi A, and Gomez-Lopez N. 2019. Are B cells altered in the decidua of women with preterm or term labor? Am J Reprod Immunol 81: e13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller D, Gershater M, Slutsky R, Romero R, and Gomez-Lopez N. 2020. Maternal and fetal T cells in term pregnancy and preterm labor. Cell Mol Immunol 17: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arenas-Hernandez M, Gomez-Lopez N, Garcia-Flores V, Rangel-Escareno C, Alvarez-Salas LM, Martinez-Acuna N, Vazquez-Perez JA, and Vega-Sanchez R. 2019. Choriodecidual leukocytes display a unique gene expression signature in spontaneous labor at term. Genes Immun 20: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, and Arenas-Hernandez M. 2014. Immune cells in term and preterm labor. Cell Mol Immunol 11: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St Louis D, Romero R, Plazyo O, Arenas-Hernandez M, Panaitescu B, Xu Y, Milovic T, Xu Z, Bhatti G, Mi QS, Drewlo S, Tarca AL, Hassan SS, and Gomez-Lopez N. 2016. Invariant NKT Cell Activation Induces Late Preterm Birth That Is Attenuated by Rosiglitazone. J Immunol 196: 1044–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Romero R, Miller D, Kadam L, Mial TN, Plazyo O, Garcia-Flores V, Hassan SS, Xu Z, Tarca AL, Drewlo S, and Gomez-Lopez N. 2016. An M1-like Macrophage Polarization in Decidual Tissue during Spontaneous Preterm Labor That Is Attenuated by Rosiglitazone Treatment. J Immunol 196: 2476–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arenas-Hernandez M, Romero R, Xu Y, Panaitescu B, Garcia-Flores V, Miller D, Ahn H, Done B, Hassan SS, Hsu CD, Tarca AL, Sanchez-Torres C, and Gomez-Lopez N. 2019. Effector and Activated T Cells Induce Preterm Labor and Birth That Is Prevented by Treatment with Progesterone. J Immunol 202: 2585–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slutsky R, Romero R, Xu Y, Galaz J, Miller D, Done B, Tarca AL, Gregor S, Hassan SS, Leng Y, and Gomez-Lopez N. 2019. Exhausted and Senescent T Cells at the Maternal-Fetal Interface in Preterm and Term Labor. J Immunol Res 2019: 3128010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, and Surani MA. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382. [DOI] [PubMed] [Google Scholar]
- 53.Hashimshony T, Wagner F, Sher N, and Yanai I. 2012. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2: 666–673. [DOI] [PubMed] [Google Scholar]
- 54.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, and Sandberg R. 2012. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, and Amit I. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343: 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161: 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, and McCarroll SA. 2015. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161: 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mora-Castilla S, To C, Vaezeslami S, Morey R, Srinivasan S, Dumdie JN, Cook-Andersen H, Jenkins J, and Laurent LC. 2016. Miniaturization Technologies for Efficient Single-Cell Library Preparation for Next-Generation Sequencing. J Lab Autom 21: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, and Bielas JH. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aldridge S, and Teichmann SA. 2020. Single cell transcriptomics comes of age. Nat Commun 11: 4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez J, Ong IM, and Stanic AK. 2019. Single-cell technologies in reproductive immunology. Am J Reprod Immunol 82: e13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Huang Q, Liu Y, and Garmire LX. 2020. Single cell transcriptome research in human placenta. Reproduction 160: R155–r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krjutškov K, Katayama S, Saare M, Vera-Rodriguez M, Lubenets D, Samuel K, Laisk-Podar T, Teder H, Einarsdottir E, Salumets A, and Kere J. 2016. Single-cell transcriptome analysis of endometrial tissue. Hum Reprod 31: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, Pan W, Simon C, and Quake SR. 2020. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 26: 1644–1653. [DOI] [PubMed] [Google Scholar]
- 65.Vilella F, and Simon C. 2021. Reproductive medicine, as seen through single-cell glasses. Fertil Steril 115: 296–297. [DOI] [PubMed] [Google Scholar]
- 66.Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, Lin HY, Zhu C, Ge H, Cross JC, and Wang H. 2018. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res 28: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, and Williams Z. 2018. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv 4: eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, and Teichmann SA. 2018. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun T, Gonzalez TL, Deng N, DiPentino R, Clark EL, Lee B, Tang J, Wang Y, Stripp BR, Yao C, Tseng HR, Karumanchi SA, Koeppel AF, Turner SD, Farber CR, Rich SS, Wang ET, Williams J, and Pisarska MD. 2020. Sexually Dimorphic Crosstalk at the Maternal-Fetal Interface. J Clin Endocrinol Metab 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi Y, Zhu H, Klausen C, and Leung PCK. 2021. Transcription factor SOX4 facilitates BMP2-regulated gene expression during invasive trophoblast differentiation. FASEB J 35: e22028. [DOI] [PubMed] [Google Scholar]
- 71.Du L, Deng W, Zeng S, Xu P, Huang L, Liang Y, Wang Y, Xu H, Tang J, Bi S, Zhang L, Li Y, Ren L, Lin L, Deng W, Liu M, Chen J, Wang H, and Chen D. 2021. Single-cell transcriptome analysis reveals defective decidua stromal niche attributes to recurrent spontaneous abortion. Cell Prolif 54: e13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He JP, Tian Q, Zhu QY, and Liu JL. 2021. Identification of Intercellular Crosstalk between Decidual Cells and Niche Cells in Mice. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang F, Jia W, Fan M, Shao X, Li Z, Liu Y, Ma Y, Li YX, Li R, Tu Q, and Wang YL. 2021. Single-cell Immune Landscape of Human Recurrent Miscarriage. Genomics Proteomics Bioinformatics 19: 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo C, Cai P, Jin L, Sha Q, Yu Q, Zhang W, Jiang C, Liu Q, Zong D, Li K, Fang J, Lu F, Wang Y, Li D, Lin J, Li L, Zeng Z, Tong X, Wei H, and Qu K. 2021. Single-cell profiling of the human decidual immune microenvironment in patients with recurrent pregnancy loss. Cell Discov 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen P, Zhou L, Chen J, Lu Y, Cao C, Lv S, Wei Z, Wang L, Chen J, Hu X, Wu Z, Zhou X, Su D, Deng X, Zeng C, Wang H, Pu Z, Diao R, and Mou L. 2021. The Immune Atlas of Human Deciduas With Unexplained Recurrent Pregnancy Loss. Front Immunol 12: 689019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shannon MJ, Baltayeva J, Castellana B, Wachter J, McNeill GL, Yoon JS, Treissman J, Le HT, Lavoie PM, and Beristain AG. 2022. Cell trajectory modeling identifies a primitive trophoblast state defined by BCAM enrichment. Development 149. [DOI] [PubMed] [Google Scholar]
- 77.Ma W, Cao M, Bi S, Du L, Chen J, Wang H, Jiang Y, Wu Y, Liao Y, Kong S, and Liu J. 2022. MAX deficiency impairs human endometrial decidualization through down-regulating OSR2 in women with recurrent spontaneous abortion. Cell Tissue Res 388: 453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pavlicev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, and Jones H. 2017. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res 27: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni YB, To KF, Cheng YKY, Chiu RWK, and Lo YMD. 2017. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci U S A 114: E7786–E7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, Leng Y, Luca F, Hassan SS, and Gomez-Lopez N. 2019. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang G, Zhou C, Jiang X, Zhang Y, Huang B, Gao S, Kang Z, Ma D, Wang F, Gottgens B, Wang H, Han JJ, and Liu F. 2021. De novo generation of macrophage from placenta-derived hemogenic endothelium. Dev Cell 56: 2121–2133 e2126. [DOI] [PubMed] [Google Scholar]
- 82.Marsh B, and Blelloch R. 2020. Single nuclei RNA-seq of mouse placental labyrinth development. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nelson AC, Mould AW, Bikoff EK, and Robertson EJ. 2016. Single-cell RNA-seq reveals cell type-specific transcriptional signatures at the maternal-foetal interface during pregnancy. Nat Commun 7: 11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toothaker JM, Olaloye O, McCourt BT, McCourt CC, Silva TN, Case RM, Liu P, Yimlamai D, Tseng G, and Konnikova L. 2022. Immune landscape of human placental villi using single-cell analysis. Development 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou X, Xu Y, Ren S, Liu D, Yang N, Han Q, Kong S, Wang H, Deng W, Qi H, and Lu J. 2021. Single-cell RNA-seq revealed diverse cell types in the mouse placenta at mid-gestation. Exp Cell Res 405: 112715. [DOI] [PubMed] [Google Scholar]
- 86.Huang JR, Nie J, Liu LJ, Zhang XW, Xie YM, Peng QZ, Wang WN, Pei CL, Zhao YH, Liu R, Huang LH, Li TP, Xie MK, and Zhang WS. 2020. Single-cell transcriptomics reveals the heterogeneity of the decidual endothelial cells that participate in labor onset. Eur Rev Med Pharmacol Sci 24: 10359–10365. [DOI] [PubMed] [Google Scholar]
- 87.Huang J, Li Q, Peng Q, Xie Y, Wang W, Pei C, Zhao Y, Liu R, Huang L, Li T, Xie L, Zhang J, Dai L, Chen J, Sun J, and Zhang W. 2021. Single-cell RNA sequencing reveals heterogeneity and differential expression of decidual tissues during the peripartum period. Cell Prolif 54: e12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang J, Zhang W, Zhao Y, Li J, Xie M, Lu Y, Peng Q, Zhang J, Li P, and Dai L. 2021. Deciphering the Intercellular Communication Network of Peripartum Decidua that Orchestrates Delivery. Front Cell Dev Biol 9: 770621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pique-Regi R, Romero R, Garcia-Flores V, Peyvandipour A, Tarca AL, Pusod E, Galaz J, Miller D, Bhatti G, Para R, Kanninen T, Hadaya O, Paredes C, Motomura K, Johnson JR, Jung E, Hsu CD, Berry SM, and Gomez-Lopez N. 2022. A single-cell atlas of the myometrium in human parturition. JCI Insight 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia-Flores V, Romero R, Peyvandipour A, Galaz J, Pusod E, Panaitescu B, Miller D, Xu Y, Tao L, Liu Y, Tarca AL, Pique-Regi R, and Gomez-Lopez N. 2022. The single-cell atlas of the murine reproductive tissues during preterm labor. bioRxiv: 2022.2004.2027.489704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pique-Regi R, Romero R, Tarca AL, Luca F, Xu Y, Alazizi A, Leng Y, Hsu CD, and Gomez-Lopez N. 2020. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li M, Chen L, Zhang J, Xiong C, and Li X. 2020. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS One 15: e0230295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu-Culligan A, Chavan AR, Vijayakumar P, Irshaid L, Courchaine EM, Milano KM, Tang Z, Pope SD, Song E, Vogels CBF, Lu-Culligan WJ, Campbell KH, Casanovas-Massana A, Bermejo S, Toothaker JM, Lee HJ, Liu F, Schulz W, Fournier J, Muenker MC, Moore AJ, Yale IT, Konnikova L, Neugebauer KM, Ring A, Grubaugh ND, Ko AI, Morotti R, Guller S, Kliman HJ, Iwasaki A, and Farhadian SF. 2021. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med (N Y) 2: 591–610 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Constantino FB, Cury SS, Nogueira CR, Carvalho RF, and Justulin LA. 2021. Prediction of Non-canonical Routes for SARS-CoV-2 Infection in Human Placenta Cells. Front Mol Biosci 8: 614728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cui D, Liu Y, Jiang X, Ding C, Poon LC, Wang H, and Yang H. 2021. Single-cell RNA expression profiling of SARS-CoV-2-related ACE2 and TMPRSS2 in human trophectoderm and placenta. Ultrasound Obstet Gynecol 57: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Q, Wang W, Pei C, Zhao Y, Liu R, Zhang W, Huang L, Li T, and Huang J. 2021. Expression of SARS-CoV-2 entry genes ACE2 and TMPRSS2 at single cell resolution in the peripartum decidua. Am J Transl Res 13: 4389–4400. [PMC free article] [PubMed] [Google Scholar]
- 97.Garcia-Flores V, Romero R, Xu Y, Theis KR, Arenas-Hernandez M, Miller D, Peyvandipour A, Bhatti G, Galaz J, Gershater M, Levenson D, Pusod E, Tao L, Kracht D, Florova V, Leng Y, Motomura K, Para R, Faucett M, Hsu CD, Zhang G, Tarca AL, Pique-Regi R, and Gomez-Lopez N. 2022. Maternal-fetal immune responses in pregnant women infected with SARS-CoV-2. Nat Commun 13: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen J, Du L, Wang F, Shao X, Wang X, Yu W, Bi S, Chen D, Pan X, Zeng S, Huang L, Liang Y, Li Y, Chen R, Xue F, Li X, Wang S, Zhuang M, Liu M, Lin L, Yan H, He F, Yu L, Jiang Q, Xiong Z, Zhang L, Cao B, Wang YL, and Chen D. 2022. Cellular and molecular atlas of the placenta from a COVID-19 pregnant woman infected at midgestation highlights the defective impacts on foetal health. Cell Prolif 55: e13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang Z, Xia S, Mei S, Wen Y, Liu J, Dong C, Chen W, Yu P, Qu L, Luo Y, and Zheng L. 2022. Integrated Analysis Reveals the Characteristics and Effects of SARS-CoV-2 Maternal-Fetal Transmission. Front Microbiol 13: 813187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pijnenborg R, Vercruysse L, and Brosens I. 2011. Deep placentation. Best Pract Res Clin Obstet Gynaecol 25: 273–285. [DOI] [PubMed] [Google Scholar]
- 101.Burton GJ, and Jauniaux E. 2015. What is the placenta? Am J Obstet Gynecol 213: S6 e1, S6–8. [DOI] [PubMed] [Google Scholar]
- 102.Brosens I, Puttemans P, and Benagiano G. 2019. Placental bed research: I. The placental bed: from spiral arteries remodeling to the great obstetrical syndromes. Am J Obstet Gynecol 221: 437–456. [DOI] [PubMed] [Google Scholar]
- 103.Albrecht ED, and Pepe GJ. 2020. Regulation of Uterine Spiral Artery Remodeling: a Review. Reprod Sci 27: 1932–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sato Y 2020. Endovascular trophoblast and spiral artery remodeling. Mol Cell Endocrinol 503: 110699. [DOI] [PubMed] [Google Scholar]
- 105.Lu X, Wang R, Zhu C, Wang H, Lin HY, Gu Y, Cross JC, and Wang H. 2017. Fine-Tuned and Cell-Cycle-Restricted Expression of Fusogenic Protein Syncytin-2 Maintains Functional Placental Syncytia. Cell Rep 21: 1150–1159. [DOI] [PubMed] [Google Scholar]
- 106.Haider S, Meinhardt G, Saleh L, Fiala C, Pollheimer J, and Knofler M. 2016. Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc Natl Acad Sci U S A 113: E7710–E7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saelens W, Cannoodt R, Todorov H, and Saeys Y. 2019. A comparison of single-cell trajectory inference methods. Nat Biotechnol 37: 547–554. [DOI] [PubMed] [Google Scholar]
- 108.Henry VJ, Bandrowski AE, Pepin AS, Gonzalez BJ, and Desfeux A. 2014. OMICtools: an informative directory for multi-omic data analysis. Database (Oxford) 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zappia L, Phipson B, and Oshlack A. 2018. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput Biol 14: e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Farley AE, Graham CH, and Smith GN. 2004. Contractile properties of human placental anchoring villi. Am J Physiol Regul Integr Comp Physiol 287: R680–685. [DOI] [PubMed] [Google Scholar]
- 111.Thornburg KL, and Marshall N. 2015. The placenta is the center of the chronic disease universe. Am J Obstet Gynecol 213: S14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Burton GJ, and Jauniaux E. 2015. What is the placenta? Am J Obstet Gynecol 213: S6.e1, S6–8. [DOI] [PubMed] [Google Scholar]
- 113.Krop J, van der Zwan A, Ijsselsteijn ME, Kapsenberg H, Luk SJ, Hendriks SH, van der Keur C, Verleng LJ, Somarakis A, van der Meeren L, Haasnoot G, Bos M, de Miranda N, Chuva de Sousa Lopes SM, van der Hoorn MP, Koning F, Claas FHJ, Heidt S, and Eikmans M. 2022. Imaging mass cytometry reveals the prominent role of myeloid cells at the maternal-fetal interface. iScience 25: 104648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li R, Wang TY, Xu X, Emery O, Yi M, Wu SP, and DeMayo FJ. 2022. Spatial Transcriptomic Profiles of Mouse Uterine Microenvironments at Pregnancy Day 7.5. Biol Reprod. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Y, Wang R, Zhang S, Song S, Jiang C, Han G, Wang M, Ajani J, Futreal A, and Wang L. 2019. iTALK: an R Package to Characterize and Illustrate Intercellular Communication. bioRxiv: 507871. [Google Scholar]
- 116.Browaeys R, Saelens W, and Saeys Y. 2020. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 17: 159–162. [DOI] [PubMed] [Google Scholar]
- 117.Cabello-Aguilar S, Alame M, Kon-Sun-Tack F, Fau C, Lacroix M, and Colinge J. 2020. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res 48: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Efremova M, Vento-Tormo M, Teichmann SA, and Vento-Tormo R. 2020. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 15: 1484–1506. [DOI] [PubMed] [Google Scholar]
- 119.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q. 2021. Inference and analysis of cell-cell communication using CellChat. Nat Commun 12: 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, Claas FH, Walker JJ, Redman CW, Morgan L, Tower C, Regan L, Moore GE, Carrington M, and Moffett A. 2010. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J Clin Invest 120: 4102–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Colucci F 2017. The role of KIR and HLA interactions in pregnancy complications. Immunogenetics 69: 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huhn O, Chazara O, Ivarsson MA, Retière C, Venkatesan TC, Norman PJ, Hilton HG, Jayaraman J, Traherne JA, Trowsdale J, Ito M, Kling C, Parham P, Ghadially H, Moffett A, Sharkey AM, and Colucci F. 2018. High-Resolution Genetic and Phenotypic Analysis of KIR2DL1 Alleles and Their Association with Pre-Eclampsia. J Immunol 201: 2593–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johnsen GM, Størvold GL, Drabbels JJM, Haasnoot GW, Eikmans M, Spruyt-Gerritse MJ, Alnæs-Katjavivi P, Scherjon SA, Redman CWG, Claas FHJ, and Staff AC. 2018. The combination of maternal KIR-B and fetal HLA-C2 is associated with decidua basalis acute atherosis in pregnancies with preeclampsia. J Reprod Immunol 129: 23–29. [DOI] [PubMed] [Google Scholar]
- 124.Repnik U, Tilburgs T, Roelen DL, van der Mast BJ, Kanhai HH, Scherjon S, and Claas FH. 2008. Comparison of macrophage phenotype between decidua basalis and decidua parietalis by flow cytometry. Placenta 29: 405–412. [DOI] [PubMed] [Google Scholar]
- 125.Kwan M, Hazan A, Zhang J, Jones RL, Harris LK, Whittle W, Keating S, Dunk CE, and Lye SJ. 2014. Dynamic changes in maternal decidual leukocyte populations from first to second trimester gestation. Placenta 35: 1027–1034. [DOI] [PubMed] [Google Scholar]
- 126.Shah NM, Herasimtschuk AA, Boasso A, Benlahrech A, Fuchs D, Imami N, and Johnson MR. 2017. Changes in T Cell and Dendritic Cell Phenotype from Mid to Late Pregnancy Are Indicative of a Shift from Immune Tolerance to Immune Activation. Front Immunol 8: 1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van der Zwan A, van Unen V, Beyrend G, Laban S, van der Keur C, Kapsenberg HJM, Höllt T, Chuva de Sousa Lopes SM, van der Hoorn MP, Koning F, Claas FHJ, Eikmans M, and Heidt S. 2020. Visualizing Dynamic Changes at the Maternal-Fetal Interface Throughout Human Pregnancy by Mass Cytometry. Front Immunol 11: 571300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marsh B, Zhou Y, Kapidzic M, Fisher S, and Blelloch R. 2022. Regionally distinct trophoblast regulate barrier function and invasion in the human placenta. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cunningham FG, Leveno KJ, Dashe JS, Hoffman BL, Spong CY, and Casey BM. 2022. Implantation and Placental Development. In Williams Obstetrics, 26e. McGraw Hill, New York, NY. [Google Scholar]
- 130.Gomez-Lopez N, Romero R, Hassan SS, Bhatti G, Berry SM, Kusanovic JP, Pacora P, and Tarca AL. 2019. The Cellular Transcriptome in the Maternal Circulation During Normal Pregnancy: A Longitudinal Study. Front Immunol 10: 2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tarca AL, Romero R, Xu Z, Gomez-Lopez N, Erez O, Hsu CD, Hassan SS, and Carey VJ. 2019. Targeted expression profiling by RNA-Seq improves detection of cellular dynamics during pregnancy and identifies a role for T cells in term parturition. Sci Rep 9: 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tarca AL, Romero R, Pique-Regi R, Pacora P, Done B, Kacerovsky M, Bhatti G, Jaiman S, Hassan SS, Hsu CD, and Gomez-Lopez N. 2020. Amniotic fluid cell-free transcriptome: a glimpse into fetal development and placental cellular dynamics during normal pregnancy. BMC Med Genomics 13: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, and Wainscoat JS. 1997. Presence of fetal DNA in maternal plasma and serum. Lancet 350: 485–487. [DOI] [PubMed] [Google Scholar]
- 134.Lo YM, Lau TK, Zhang J, Leung TN, Chang AM, Hjelm NM, Elmes RS, and Bianchi DW. 1999. Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21. Clin Chem 45: 1747–1751. [PubMed] [Google Scholar]
- 135.Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, and Lee TH. 2001. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion 41: 1524–1530. [DOI] [PubMed] [Google Scholar]
- 136.Bianchi DW, and Lo YM. 2001. Fetomaternal cellular and plasma DNA trafficking: the Yin and the Yang. Ann N Y Acad Sci 945: 119–131. [DOI] [PubMed] [Google Scholar]
- 137.Ng EK, Tsui NB, Lau TK, Leung TN, Chiu RW, Panesar NS, Lit LC, Chan KW, and Lo YM. 2003. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci U S A 100: 4748–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chiu RW, Lui WB, Cheung MC, Kumta N, Farina A, Banzola I, Grotti S, Rizzo N, Haines CJ, and Lo YM. 2006. Time profile of appearance and disappearance of circulating placenta-derived mRNA in maternal plasma. Clin Chem 52: 313–316. [DOI] [PubMed] [Google Scholar]
- 139.Taglauer ES, Wilkins-Haug L, and Bianchi DW. 2014. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 35 Suppl: S64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yeganeh Kazemi N, Fedyshyn B, Sutor S, Fedyshyn Y, Markovic S, and Enninga EAL. 2021. Maternal Monocytes Respond to Cell-Free Fetal DNA and Initiate Key Processes of Human Parturition. J Immunol 207: 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gomez-Lopez N, Guilbert LJ, and Olson DM. 2010. Invasion of the leukocytes into the fetal-maternal interface during pregnancy. J Leukoc Biol 88: 625–633. [DOI] [PubMed] [Google Scholar]
- 142.Li J, Wang Q, An Y, Chen X, Xing Y, Deng Q, Li Z, Wang S, Dai X, Liang N, Hou Y, Yang H, and Shang Z. 2022. Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Mesenchymal Stem/Stromal Cells Derived from Human Placenta. Front Cell Dev Biol 10: 836887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miller D, Motomura K, Garcia-Flores V, Romero R, and Gomez-Lopez N. 2018. Innate Lymphoid Cells in the Maternal and Fetal Compartments. Front Immunol 9: 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Osman I, Young A, Ledingham MA, Thomson AJ, Jordan F, Greer IA, and Norman JE. 2003. Leukocyte density and pro-inflammatory cytokine expression in human fetal membranes, decidua, cervix and myometrium before and during labour at term. Mol Hum Reprod 9: 41–45. [DOI] [PubMed] [Google Scholar]
- 145.Hamilton S, Oomomian Y, Stephen G, Shynlova O, Tower CL, Garrod A, Lye SJ, and Jones RL. 2012. Macrophages infiltrate the human and rat decidua during term and preterm labor: evidence that decidual inflammation precedes labor. Biol Reprod 86: 39. [DOI] [PubMed] [Google Scholar]
- 146.Hamilton SA, Tower CL, and Jones RL. 2013. Identification of chemokines associated with the recruitment of decidual leukocytes in human labour: potential novel targets for preterm labour. PLoS One 8: e56946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shynlova O, Nedd-Roderique T, Li Y, Dorogin A, Nguyen T, and Lye SJ. 2013. Infiltration of myeloid cells into decidua is a critical early event in the labour cascade and post-partum uterine remodelling. J Cell Mol Med 17: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Provine NM, and Klenerman P. 2020. MAIT Cells in Health and Disease. Annu Rev Immunol 38: 203–228. [DOI] [PubMed] [Google Scholar]
- 149.Solders M, Gorchs L, Erkers T, Lundell AC, Nava S, Gidlöf S, Tiblad E, Magalhaes I, and Kaipe H. 2017. MAIT cells accumulate in placental intervillous space and display a highly cytotoxic phenotype upon bacterial stimulation. Sci Rep 7: 6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Solders M, Gorchs L, Gidlöf S, Tiblad E, Lundell AC, and Kaipe H. 2017. Maternal Adaptive Immune Cells in Decidua Parietalis Display a More Activated and Coinhibitory Phenotype Compared to Decidua Basalis. Stem Cells Int 2017: 8010961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Solders M, Gorchs L, Tiblad E, Gidlöf S, Leeansyah E, Dias J, Sandberg JK, Magalhaes I, Lundell AC, and Kaipe H. 2019. Recruitment of MAIT Cells to the Intervillous Space of the Placenta by Placenta-Derived Chemokines. Front Immunol 10: 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vazquez J, Chavarria M, Chasman DA, Welch Schwartz R, Tyler CT, Lopez G, Fisher RC, Ong IM, and Stanic AK. 2021. Multiomic analysis reveals decidual-specific transcriptional programing of MAIT cells. Am J Reprod Immunol 86: e13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Romero R, Dey SK, and Fisher SJ. 2014. Preterm labor: one syndrome, many causes. Science 345: 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jung E, Romero R, Yeo L, Gomez-Lopez N, Chaemsaithong P, Jaovisidha A, Gotsch F, and Erez O. 2022. The etiology of preeclampsia. Am J Obstet Gynecol 226: S844–S866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang T, Bian Q, Chen Y, Wang X, Yu S, Liu S, Ji P, Li L, Shrestha M, Dong S, Guo R, and Zhang H. 2021. Dissecting human trophoblast cell transcriptional heterogeneity in preeclampsia using single-cell RNA sequencing. Mol Genet Genomic Med 9: e1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Guo F, Zhang B, Yang H, Fu Y, Wang Y, Huang J, Cheng M, Li X, Shen Z, Li L, He P, Xiang AP, Wang S, and Zhang H. 2021. Systemic transcriptome comparison between early- And late-onset pre-eclampsia shows distinct pathology and novel biomarkers. Cell Prolif 54: e12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou W, Wang H, Yang Y, Guo F, Yu B, and Su Z. 2022. Trophoblast Cell Subtypes and Dysfunction in the Placenta of Individuals with Preeclampsia Revealed by SingleCell RNA Sequencing. Mol Cells 45: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.DiFederico E, Genbacev O, and Fisher SJ. 1999. Preeclampsia is associated with widespread apoptosis of placental cytotrophoblasts within the uterine wall. Am J Pathol 155: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Leung DN, Smith SC, To KF, Sahota DS, and Baker PN. 2001. Increased placental apoptosis in pregnancies complicated by preeclampsia. Am J Obstet Gynecol 184: 1249–1250. [DOI] [PubMed] [Google Scholar]
- 160.Ishihara N, Matsuo H, Murakoshi H, Laoag-Fernandez JB, Samoto T, and Maruo T. 2002. Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardation. Am J Obstet Gynecol 186: 158–166. [DOI] [PubMed] [Google Scholar]
- 161.Kadyrov M, Kingdom JC, and Huppertz B. 2006. Divergent trophoblast invasion and apoptosis in placental bed spiral arteries from pregnancies complicated by maternal anemia and early-onset preeclampsia/intrauterine growth restriction. Am J Obstet Gynecol 194: 557–563. [DOI] [PubMed] [Google Scholar]
- 162.Longtine MS, Chen B, Odibo AO, Zhong Y, and Nelson DM. 2012. Villous trophoblast apoptosis is elevated and restricted to cytotrophoblasts in pregnancies complicated by preeclampsia, IUGR, or preeclampsia with IUGR. Placenta 33: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.von Dadelszen P, Magee LA, and Roberts JM. 2003. Subclassification of preeclampsia. Hypertens Pregnancy 22: 143–148. [DOI] [PubMed] [Google Scholar]
- 164.Tranquilli AL, Brown MA, Zeeman GG, Dekker G, and Sibai BM. 2013. The definition of severe and early-onset preeclampsia. Statements from the International Society for the Study of Hypertension in Pregnancy (ISSHP). Pregnancy Hypertens 3: 44–47. [DOI] [PubMed] [Google Scholar]
- 165.Miller D, Motomura K, Galaz J, Gershater M, Lee ED, Romero R, and Gomez-Lopez N. 2022. Cellular immune responses in the pathophysiology of preeclampsia. J Leukoc Biol 111: 237–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chaiworapongsa T, Chaemsaithong P, Yeo L, and Romero R. 2014. Pre-eclampsia part 1: current understanding of its pathophysiology. Nat Rev Nephrol 10: 466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Burton GJ, Redman CW, Roberts JM, and Moffett A. 2019. Pre-eclampsia: pathophysiology and clinical implications. Bmj 366: l2381. [DOI] [PubMed] [Google Scholar]
- 168.Staff AC, Fjeldstad HE, Fosheim IK, Moe K, Turowski G, Johnsen GM, Alnaes-Katjavivi P, and Sugulle M. 2022. Failure of physiological transformation and spiral artery atherosis: their roles in preeclampsia. Am J Obstet Gynecol 226: S895–s906. [DOI] [PubMed] [Google Scholar]
- 169.Norwitz ER, Robinson JN, and Challis JR. 1999. The control of labor. N Engl J Med 341: 660–666. [DOI] [PubMed] [Google Scholar]
- 170.Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, Chaiworapongsa T, and Mazor M. 2006. The preterm parturition syndrome. Bjog 113 Suppl 3: 17–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Smith R 2007. Parturition. N Engl J Med 356: 271–283. [DOI] [PubMed] [Google Scholar]
- 172.Koh W, Wu A, Penland L, Treutlein B, Neff NF, Mantalas GL, Blumenfeld YJ, El-Sayed YY, Stevenson DK, Shaw GM, and Quake SR. 2019. Single Cell Transcriptomes Derived from Human Cervical and Uterine Tissue during Pregnancy. Adv Biosyst 3: e1800336. [DOI] [PubMed] [Google Scholar]
- 173.Blum JS, Wearsch PA, and Cresswell P. 2013. Pathways of antigen processing. Annu Rev Immunol 31: 443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Farr L, Ghosh S, Jiang N, Watanabe K, Parlak M, Bucala R, and Moonah S. 2020. CD74 Signaling Links Inflammation to Intestinal Epithelial Cell Regeneration and Promotes Mucosal Healing. Cell Mol Gastroenterol Hepatol 10: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rodor J, Chen SH, Scanlon JP, Monteiro JP, Caudrillier A, Sweta S, Stewart KR, Shmakova A, Dobie R, Henderson BEP, Stewart K, Hadoke PWF, Southwood M, Moore SD, Upton PD, Morrell NW, Li Z, Chan SY, Handen A, Lafyatis R, de Rooij L, Henderson NC, Carmeliet P, Spiroski AM, Brittan M, and Baker AH. 2021. Single-cell RNA-seq profiling of mouse endothelial cells in response to pulmonary arterial hypertension. Cardiovasc Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Romero R, Brody DT, Oyarzun E, Mazor M, Wu YK, Hobbins JC, and Durum SK. 1989. Infection and labor. III. Interleukin-1: a signal for the onset of parturition. Am J Obstet Gynecol 160: 1117–1123. [DOI] [PubMed] [Google Scholar]
- 177.Romero R, Durum S, Dinarello CA, Oyarzun E, Hobbins JC, and Mitchell MD. 1989. Interleukin-1 stimulates prostaglandin biosynthesis by human amnion. Prostaglandins 37: 13–22. [DOI] [PubMed] [Google Scholar]
- 178.Bry K, and Hallman M. 1991. Synergistic stimulation of amnion cell prostaglandin E2 synthesis by interleukin-1, tumor necrosis factor and products from activated human granulocytes. Prostaglandins Leukot Essent Fatty Acids 44: 241–245. [DOI] [PubMed] [Google Scholar]
- 179.Romero R, Mazor M, Brandt F, Sepulveda W, Avila C, Cotton DB, and Dinarello CA. 1992. Interleukin-1 alpha and interleukin-1 beta in preterm and term human parturition. Am J Reprod Immunol 27: 117–123. [DOI] [PubMed] [Google Scholar]
- 180.Wienke J, Brouwers L, van der Burg LM, Mokry M, Scholman RC, Nikkels PG, van Rijn BB, and van Wijk F. 2020. Human Tregs at the materno-fetal interface show site-specific adaptation reminiscent of tumor Tregs. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, and Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581: 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, and Qi J. 2020. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 181: 894–904 e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, and Pohlmann S. 2020. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181: 271–280 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zheng QL, Duan T, and Jin LP. 2020. Single-cell RNA expression profiling of ACE2 and AXL in the human maternal-Fetal interface. Reprod Dev Med 4: 7–10. [Google Scholar]
- 185.Ashary N, Bhide A, Chakraborty P, Colaco S, Mishra A, Chhabria K, Jolly MK, and Modi D. 2020. Single-Cell RNA-seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors Driving Pathogenesis of SARS-CoV-2. Front Cell Dev Biol 8: 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Beesley MA, Davidson JR, Panariello F, Shibuya S, Scaglioni D, Jones BC, Maksym K, Ogunbiyi O, Sebire NJ, Cacchiarelli D, David AL, De Coppi P, and Gerli M. 2022. COVID-19 and vertical transmission: assessing the expression of ACE2/TMPRSS2 in the human fetus and placenta to assess the risk of SARS-CoV-2 infection. Bjog 129: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.See P, Lum J, Chen J, and Ginhoux F. 2018. A Single-Cell Sequencing Guide for Immunologists. Front Immunol 9: 2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen G, Ning B, and Shi T. 2019. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front Genet 10: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lahnemann D, Koster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A, Pinello L, Skums P, Stamatakis A, Attolini CS, Aparicio S, Baaijens J, Balvert M, Barbanson B, Cappuccio A, Corleone G, Dutilh BE, Florescu M, Guryev V, Holmer R, Jahn K, Lobo TJ, Keizer EM, Khatri I, Kielbasa SM, Korbel JO, Kozlov AM, Kuo TH, Lelieveldt BPF, Mandoiu II, Marioni JC, Marschall T, Molder F, Niknejad A, Raczkowski L, Reinders M, Ridder J, Saliba AE, Somarakis A, Stegle O, Theis FJ, Yang H, Zelikovsky A, McHardy AC, Raphael BJ, Shah SP, and Schonhuth A. 2020. Eleven grand challenges in single-cell data science. Genome Biol 21: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kharchenko PV 2021. The triumphs and limitations of computational methods for scRNA-seq. Nat Methods 18: 723–732. [DOI] [PubMed] [Google Scholar]
- 191.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S. 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Garcia-Flores V, Xu Y, Pusod E, Romero R, Pique-Regi R, and Gomez-Lopez N. Accepted, 2022. Preparation of single-cell suspensions from the human placenta. [DOI] [PMC free article] [PubMed]
- 193.Hafemeister C, and Satija R. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. 2019. Comprehensive Integration of Single-Cell Data. Cell 177: 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, and Ye CJ. 2018. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yu G, Wang LG, Han Y, and He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]


