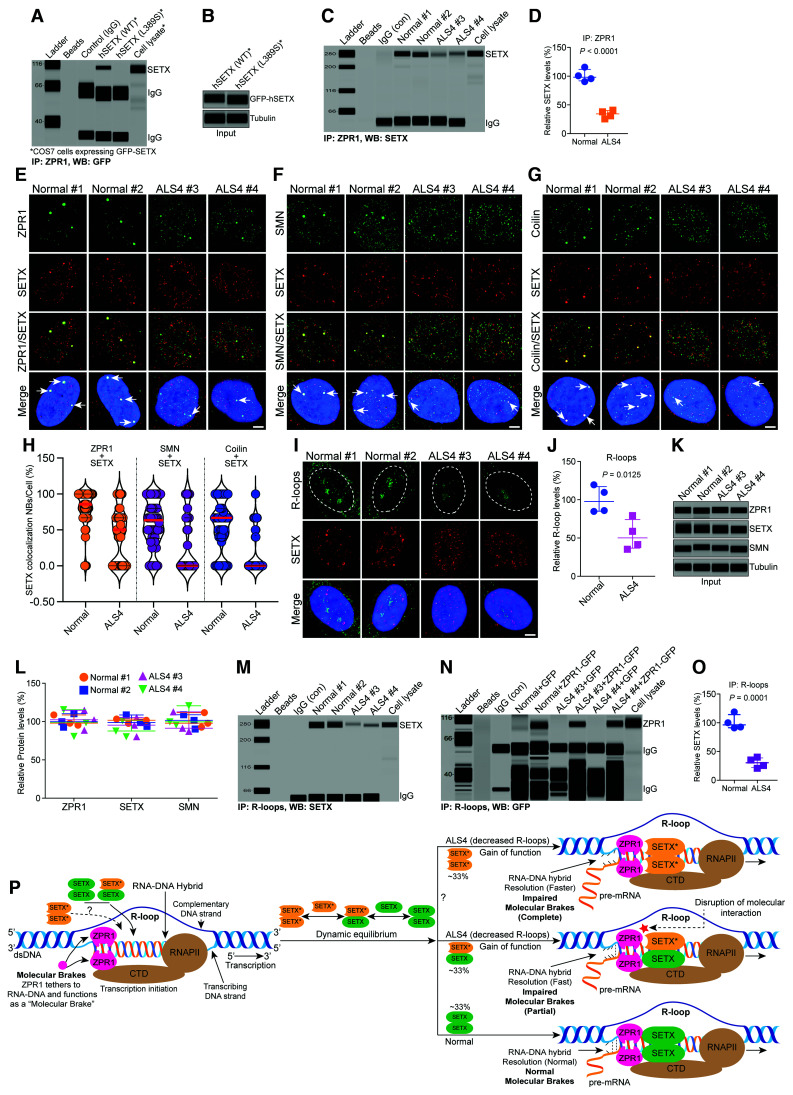
Figure 7.
Interaction of SETX with ZPR1 is disrupted in ALS4 patients and ZPR1 fails to recruit mutant SETX onto R-loops in ALS4. Mutational analysis shows that SETX L389S mutation, which causes autosomal dominant ALS4, disrupts interaction of SETX with ZPR1. (A) COS7 cells were transfected with plasmids pDEST53 expressing GFP-hSETX (1–667) (WT) and GFP-hSETX (1–667) (L389S). Immunoprecipitation was performed using anti-ZPR1 antibody followed by western blot with anti-GFP to detect GFP-hSETX. (B) Immunoblots of cell lysate from cells expressing GFP-hSETX (WT) and GFP-hSETX (L389S). To gain insight into the contribution of disruption of SETX-ZPR1 complexes in the pathogenesis of ALS4, we used fibroblasts derived from normal subjects and ALS4 patients that have heterozygous SETX mutation L389S (SETX+/L389S). Cultured control, Normal #1 and Normal #2, and ALS4 cell lines, ALS4 #3 and ALS4 #4 were used for immunofluorescence, immunoblot and immunoprecipitation analyses. (C) Interaction of SETX with ZPR1 is disrupted in ALS4 patients. Immunoprecipitation of ZPR1 shows decrease in co-immunoprecipitation of SETX from ALS4 patients compared to control (Normal) fibroblast. (D) Quantitation of SETX co-IP with ZPR1 in Normal and ALS4 patient cells is presented as a scatter plot with median and range (min, median, max). Normal (90.54, 98.21, 115.70), ALS4 (25.54, 34.44, 40.87). Quantitation of SETX levels (mean ± SEM, n = 4) in ZPR1 immunoprecipitation shows that the levels of SETX binding with ZPR1 are reduced to (33.82 ± 3.45%, P < 0.0001) in ALS4 compared to control cells. Representative images with highest co-localization are presented for double-labelled immunostainings: (E) Control cells (Normal #1 and Normal #2) stained with ZPR1 (green) and SETX (red) show ZPR1 and SETX co-localize in subnuclear bodies (arrows). In contrast, SETX mutation causes partial disruption of subnuclear bodies and mislocalization of ZPR1 and SETX in ALS4 patient cells. (F) Cells stained with SMN (green) and SETX (red) show SETX co-localizes with SMN in nuclear gems bodies (arrows) in cells from Normal subjects. The SETX mutation causes partial disruption of SMN+ gems and results in smaller SMN+ foci in ALS4 patient cells. (G) Cells stained with coilin (green) and SETX (red) show SETX co-localizes with coilin in Cajal bodies (arrows) in Normal cells. Notably, SETX mutation causes disruption of CBs and results in smaller CBs in ALS4 compared to Normal cells. (H) Quantification of SETX co-localization in subnuclear bodies (NBs)/cell (%) in normal and ALS4 patient cells is shown as a violin plot with median and interquartile range (Q1, median, Q3) (50 cells/group). In normal cells, SETX co-localization with ZPR1: ZPR1 + SETX (66.67, 85.71, 100.00), SMN: SMN + SETX (38.33, 63.33, 80.00) and coilin: Coilin + SETX (38.33, 66.67. 80.00). In ALS4 cells, SETX co-localization with ZPR1: ZPR1 + SETX (0.0, 50.00, 66.67), SMN: SMN + SETX (0.0, 0.0, 66.67) and coilin: Coilin + SETX (0.0, 0.0. 42.50). Quantification of SETX co-localization in normal cells (mean ± SEM, n = 50 cells/group) shows higher co-localization with ZPR1 (80.04 ± 3.64%) compared to SMN (57.87 ± 4.60%) (gems) and Coilin (55.06 ± 4.99%) (Cajal bodies). SETX co-localization in ALS4 cells also show higher co-localization with ZPR1 (41.88 ± 5.21%) compared to SMN (32.50 ± 5.82%) and Coilin (17.60 ± 4.17%). The overall SETX co-localization is reduced by ∼48% (ZPR1, P < 0.0001), ∼44% (SMN, P = 0.0009) and ∼68% (Coilin, P < 0.0001) in ALS4 compared to normal cells. (I) Immunofluorescence analysis of R-loops (green) and SETX (red) shows reduced intensity of R-loops in ALS4 compared to normal cells. Nuclei were stained with DAPI (blue). Scale bar = 5.0 μm. (J) Quantitation of R-loop nuclear immunofluorescence intensity in Normal and ALS4 patient cells is shown as a scatter plot with median and range. Normal (84.76, 97.82, 119.60), ALS4 (35.28, 50.17, 79.25). Quantitative (mean ± SEM, n = 4) and statistical (unpaired t-test) analysis of R-loop nuclear immunofluorescence intensity in cells show fewer R-loops (53.72 ± 9.85%, P = 0.0125) in ALS4 patient cells (ALS4 #3 + ALS4 #4) compared to control (Normal #1 + Normal #2) cells. Quantification of R-loop levels using dot-blot analysis is included in Supplementary Fig. 12. (K) Immunoblot analysis of proteins ZPR1, SETX, SMN and tubulin in Normal #1 and Normal #2, and ALS4 #3 and ALS4 #4, patient cells. (L) Quantitation of protein levels in Normal and ALS4 patient cells is shown as a scatter plot with median and range (min, median, max). ZPR1: Normal #1 (95.36, 97.98, 108.7), Normal #2 (92.65, 99.85, 110.30), and ALS4 #3 (88.65, 102.40, 114.90), ALS4 #4 (88.65, 102.40, 114.9); SETX: Normal #1 (97.65, 101.70, 103.30), Normal #2 (93.75, 98.73, 108.7), and ALS4 #3 (80.32, 95.32, 105.40), ALS4 #4 (80.25, 87.69, 101.30); SMN: Normal #1 (92.65, 97.85, 111.70), Normal #2 (90.35, 101.50, 109.00), and ALS4 #3 (87.21, 91.25, 112.70), ALS4 #4 (95.64, 99.65, 120.70). Quantitative analysis of protein levels (mean ± SEM, n = 3) shows SETX mutation did not significantly change the levels of ZPR1 (98.27 ± 10.10%, P = 0.8252), SMN (97.04 ± 7.89%, P = 0.9080) and SETX (93.67 ± 7.27%, P = 0.3903) in ALS4 compared to normal cells. (M) SETX fails to associate in vivo with R-loops in ALS4. IP of R-loops shows marked decrease in SETX co-immunoprecipitation in ALS4 compared to control suggesting that ZPR1 fails to recruit mutant SETX to R-loops. (N) Immunoprecipitations of R-loops from Normal and ALS4 subjects infected with Ad5-GFP and Ad5-ZPR1-GFP show that the binding of ZPR1 with R-loops is unaffected under ALS4 disease conditions and supports the idea that ZPR1 interaction with SETX is critical for the recruitment of SETX onto R-loops. (O) Quantitation of SETX co-IP with R-loops in Normal and ALS4 patient cells is presented as a scatter plot with median and range. Normal (91.34, 96.62, 119.2), ALS4 (20.76, 30.49, 40.32). Quantitative and statistical analysis of SETX co-IP (mean ± SEM, n = 4) with R-loop immunoprecipitation shows that SETX binding with R-loops reduced to (30.51 ± 4.43%, P = 0.0001) in ALS4 compared to control cells. (P) Graphical illustration of the molecular mechanism of R-loop resolution in ALS4 patients that have SETX L389S mutation. ZPR1 tethers to RNA:DNA hybrid and may function as a ‘molecular brake’ to control the speed of R-loop resolution by regulating translocase/helicase activity of SETX during transcription. SETX mutation alters dynamic equilibrium of SETX dimers and causes disruption of SETX-ZPR1 complexes that may result in partial impairment of the molecular brake leading to faster resolution (gain-of-function) and fewer R-loops in ALS4. All full-length blots are included in Supplementary Fig. 11.