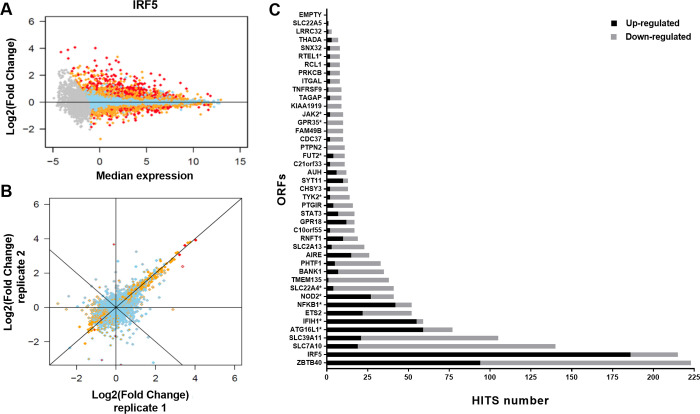
Fig 1. Impact of IBD gene candidate ORFs on the THP-1 transcriptome.
(A) Selected example illustrating impact observed on the transcriptome of THP-1 cells following the expression of IRF5. Each dot represents a single detectable gene in the THP-1 transcriptome. The x-axis shows the log2-transformed median expression across all conditions tested (baseline). The y-axis represents the effect of transduction and expression of a given ORF, as the log2-transformed fold-induction compared to baseline. Skyblue dots represent genes with expression value within expected variation (|Z|≤2), orange dots represent genes suggestively outside the range (|Z|>2) and red dots represent genes outside expected range of variation (|Z|>4). Gray dots are genes with expression value below our detection threshold. Additive effect in log2 correspond to multiplicative effect on the original scale. The fold-change equivalent to a given effect log2-effect x is then: FC = 2x. As an example, an effect of 1 correspond to a FC = 2. (B) Correlation of effect of independent sets of replicated expression of IRF5 on THP-1 transcriptome. The x-axis (inner color of dots) and y-axis (border color of dots) show the effect of two independent set of replicated ORFs on the transcriptome, as the log2-transformed fold-induction compared to baseline. Variation between sets of replicates includes effect of independent infection dates, RNA extraction, expression arrays and batches. (C) Impact of the transduction and expression of all 42 IBD gene candidate ORFs on the transcriptome of THP-1 cells. ORFs are ordered by their total number of HITS, with the number of up- and down-regulated HITS illustrated by black and gray, respectively (S2 Table & S1 Appendix). Starred ORFs are previously reported IBD candidate causal genes.