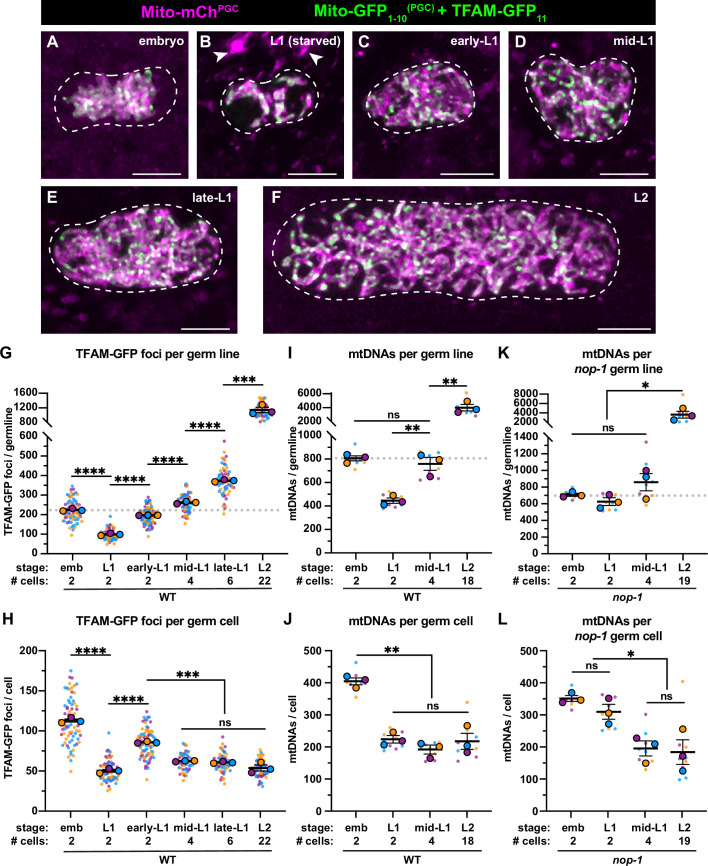
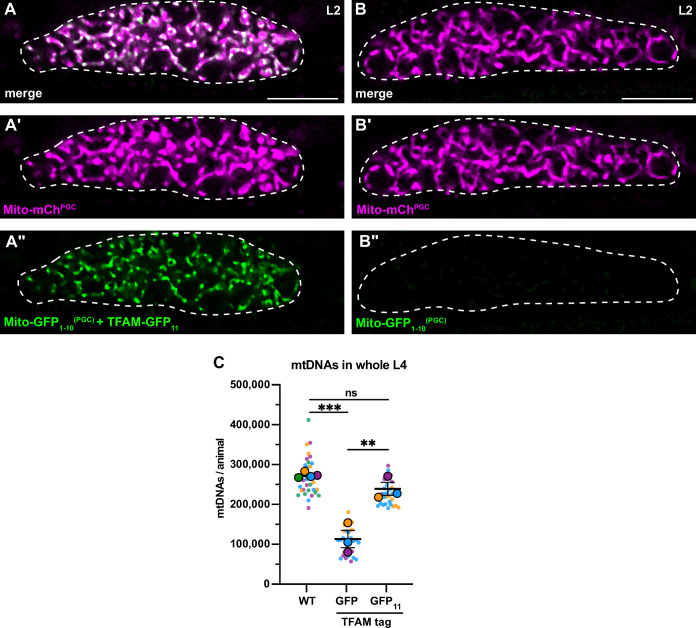
Figure 3. Primordial germ cell (PGC) lobe cannibalism and autophagy generate a mitochondrial DNA (mtDNA) low point and set point.
(A–F) Germline mitochondria and mitochondrial transcription factor-A (TFAM)-GFP11 in live embryos and larvae at the indicated stage. Dashed lines outline the PGCs or germline stem cells (GSCs). (G–H) Quantification of TFAM-GFP11 foci per germ line (G) and per germ cell (H) in embryos and larvae. (I–J) Quantification of mtDNAs per germ line (I) or per germ cell (J) in embryos and larvae; data shown for PGC mtDNA copy number in embryos and starved L1s are provided for comparison and originate from Figure 2B. (K–L) Quantification of mtDNAs per germ line (K) or per germ cell (L) in nop-1 mutant embryos and larvae; data shown for PGC mtDNA copy number in nop-1 mutant embryos and starved L1s are provided for comparison and originate from Figure 2E. Data in graphs: small dots are individual animals (TFAM-GFP11 measurements) or technical replicates (droplet digital PCR [ddPCR] experiments) from three color-coded biological replicates; the mean from each experiment is shown as a larger circle, the mean of means as a horizontal line, and the SEM as error bars. n.s., not significant (p>0.05), *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001 unpaired two-tailed Student’s t-test. Scale bars, 5 µm.