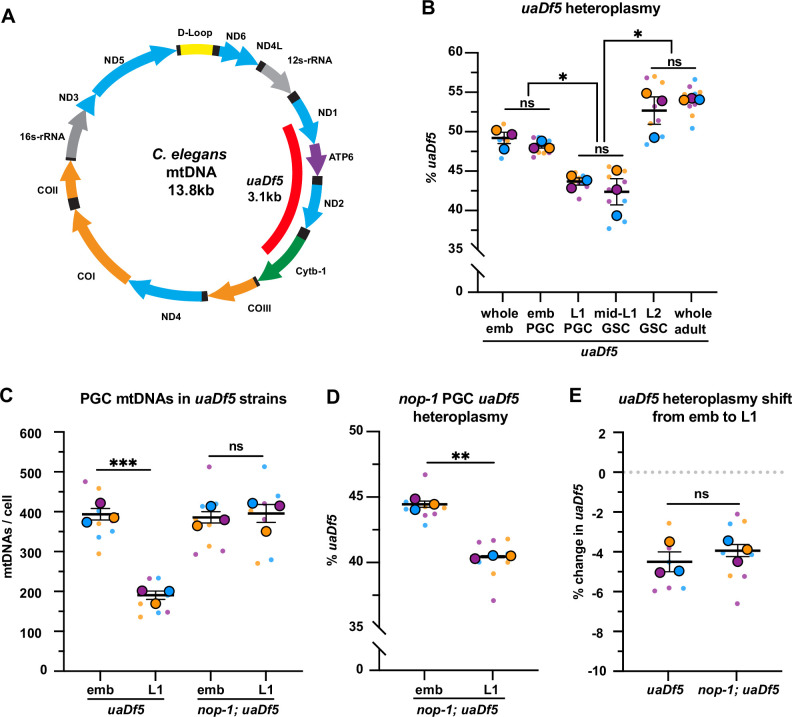
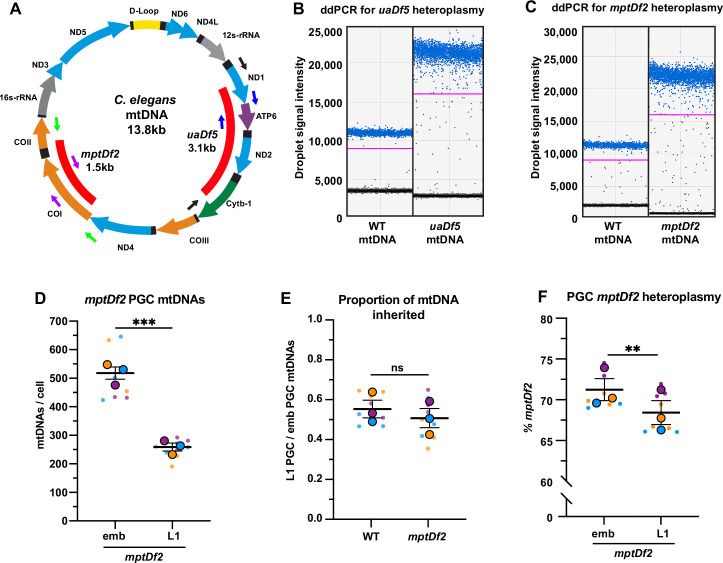
Figure 4. Primordial germ cells (PGCs) reduce uaDf5 heteroplasmy independently of lobe cannibalism.
(A) Schematic of C. elegans mitochondrial DNA (mtDNA); genes are indicated with colored arrows and the region deleted in uaDf5 is shown with a red bar. (B) Quantification of uaDf5 heteroplasmy in whole embryos, sorted PGCs or germline stem cells (GSCs), or whole adults at the indicated stages. (C) Quantification of mtDNA copy number in PGCs of uaDf5 and nop-1; uaDf5 mutants. (D) Quantification of uaDf5 heteroplasmy in nop-1; uaDf5 mutant PGCs. (E) Data from (B and D) presented as change in heteroplasmy shift from embryonic to L1 PGCs. Data in graphs: small dots are three technical replicates of droplet digital PCR (ddPCR) quantification from each of three color-coded biological replicates; the technical replicate mean from each experiment is shown as a larger circle, the mean of means as a horizontal line, and the SEM as error bars. n.s., not significant (p>0.05), *p≤0.05, **p≤0.01, ***p≤0.001, paired (B, D) and unpaired (B, C, E) two-tailed Student’s t-test.