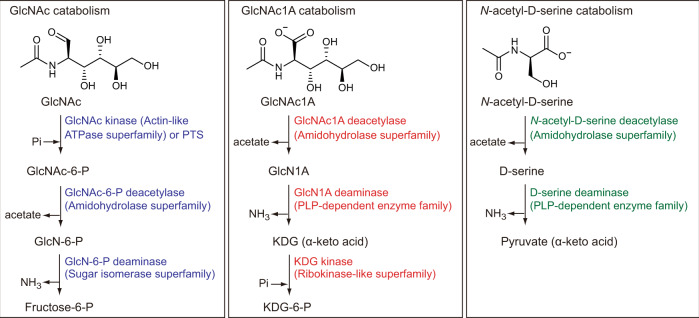
Fig. 1. The proposed catabolic pathway for GlcNAc1A in this study and reported catabolic pathways for GlcNAc and N-acetyl-D-serine.
Catabolism of GlcNAc is initiated by phosphorylation by a GlcNAc kinase (e.g. in Shewanella oneidensis11) or a PTS (e.g. in Vibrionaceae8,40), which is subsequently deacetylated and deaminated to produce fructose-6-P. Catabolism of N-acetyl-D-serine in some bacteria starts with deacetylation followed by deamination and formation of pyruvate44,54. In the proposed catabolic pathway for GlcNAc1A in this study, GlcNAc1A is deacetylated and deaminated directly to produce KDG without activation by phosphorylation. As such, this GlcNAc1A degradation pathway resembles N-acetyl-D-amino acid catabolism through deacetylation and deamination. Enzymes involved in catabolic pathways for GlcNAc1A, GlcNAc and N-acetyl-D-serine are shown in red, blue and green, respectively. The protein families of enzymes involved in each pathway are indicated in parentheses. PTS phosphotransferase, GlcN D-glucosamine, PLP pyridoxal 5-phosphate, KDG 2-keto-3-deoxygluconate.