Fig. 1. DND1 binds RNA targets mainly through its RRMs.
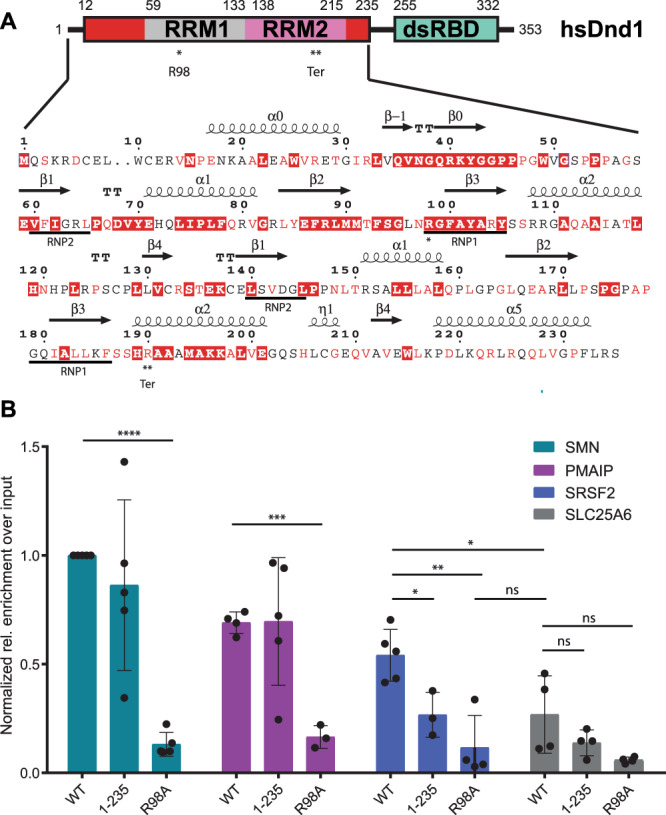
A Domain structure of DND1 and sequence of the N-terminal part of the human protein (Uniprot Q8IYX4 [https://www.uniprot.org/uniprotkb/Q8IYX4/entry]) ending at the C-terminus of the tandem RRM construct used for structure determination in this work (12–235). RRM1 in gray, RRM2 in pink, N- and C-terminal conserved extensions in red, dsRBD in green. The dsRBD-truncation mutant 1-235 used in our RIP assay ends after the extension of RRM2. Red coloring in the sequence indicates high conservation as described in Supplementary Fig. 1C. Secondary structure elements as found in our structure are indicated above the sequence. The RRM-canonical RNA-binding RNP sequences are underlined below. R98 in RNP1 that was mutated for the RIP assay is indicated with one asterisk. The Ter truncation at R190 is indicated with a double asterisk and “Ter”. See also Supplementary Fig. 1A, C,B RNA Immunoprecipitation from HEK293T cells transiently expressing FLAG-tagged DND1 or its mutants followed by qPCR using primers for published DND1 targets and negative control (Supplementary Table 1). Data from five independent experiments is presented as relative enrichment over the input (2-ΔCt), normalized to the enrichment of the SMN targets pulled down by Dnd1 WT. ΔCt is an average of (Ct [RIP] – (Ct [Input]) of technical triplicates with SD < 0.3. If SD (technical triplicate) was > 0.3 the data point was omitted. Only data with at least N = 3 is presented. The results are represented as means and SD. P values from two-tailed Welch’s t-test: *P < 0.05 (exact values 0.0191 for SRSF2 1-235 vs WT; 0.0460 for SRSF2 WT vs SLC25A6 WT); **P < 0.01 (exact value 0.0038 for SRSF2 R98A vs WT); ***P < 0.001 (exact value 0.0001 for PMAIP R98A vs WT); ****P < 0.0001. DND1 and mutants are well expressed in HEK293T cell culture (Supplementary Fig. 2). Source data are provided as a Source Data file.
