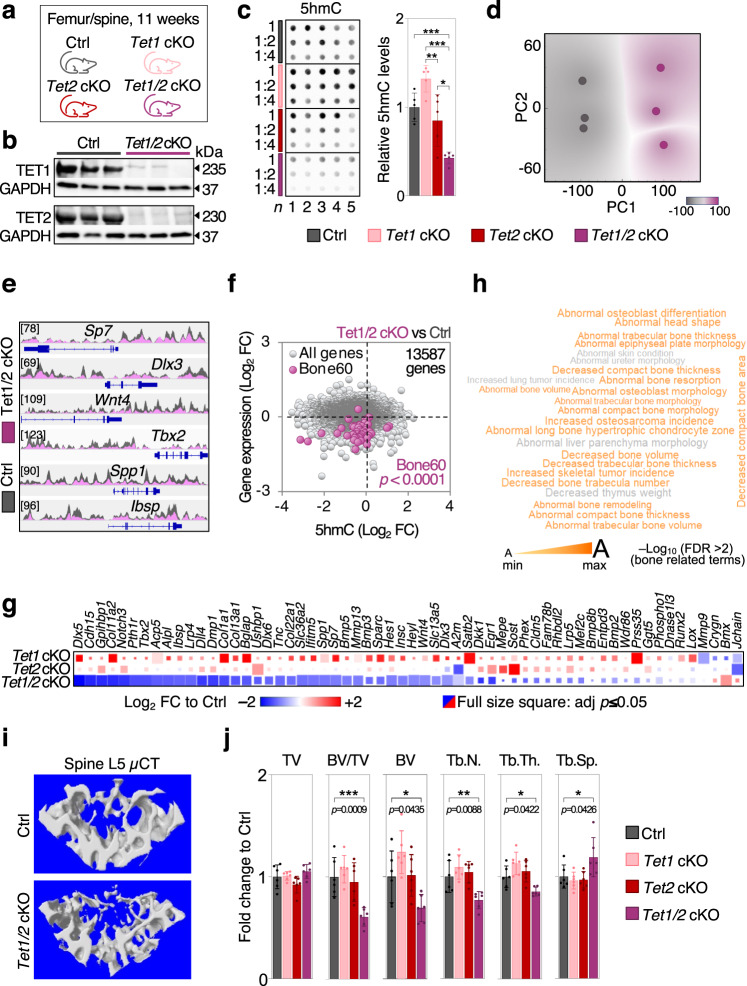
Fig. 10. Conditional Tet1 and Tet2 deletion impairs DNA hydroxymethylation at bone related loci and causes bone loss in mice.
a Schematic representation of the mouse models used. Bone tissue was collected at 11 weeks; cKO, conditional knockout. b Prrx1 driven TET1 and TET2 knockdown efficiencies in femurs of Tet1/2 double knockout mice. c 5hmC dot blot and relative quantification. d–f hMeDIP-Seq of femurs from Tet1/2 double knockout mice. d Principal component (PC) analysis and 5hmC peaks around selected bone specific genes (e). f Correlation between 5hmC occupancy (TSS + /−30 kb) and gene expression as measured by RNA-Seq; FC, fold change. g Expression of Bone60 genes in conditional Tet1, Tet2 or Tet1/2 femurs versus Ctrl femurs. h Wordcloud representing phenotypes associated with distal intergenic loci with the strongest 5hmC loss in double knockout vs Ctrl femurs as analyzed by GREAT analysis; FDR, false discovery rate. i µCT analysis of L5 spine and relative quantification; TV total volume, BV bone volume, Tb.N. trabecular number, Tb.Th. trabecular thickness, Tb.Sp. trabecular separation (j). Bar graphs represent mean ± SD; *p < 0.05; **p < 0.01; ***p < 0.001. One-way ANOVA analysis with Tukey’s (c) and Dunnett’s (j) multiple comparison tests. FDR-adjusted two-tailed, unpaired t tests in g, two-sided Fishers exact test (f). In c, Ctrl vs Tet1/2 cKO ***p = 0.0009, Tet1 cKO vs Tet1/2 cKO ***p = < 0.0001, Tet1 cKO vs Tet2 cKO **p = 0.005, Tet2 cKO vs Tet1/2 cKO *p = 0.0118. N = 5 (c) & n = 6 per group in j from biologically independent animals. Source data are provided as a Source Data File.