Abstract
Introduction
Functional single-nucleotide polymorphisms (SNPs) in genes regulating cellular uptake, elimination, and metabolism of xenobiotics may potentially influence the outcome of chronic myeloid leukemia (CML) patients treated with BCR-ABL1 tyrosine kinase inhibitors (TKI). Dasatinib, a second-generation TKI, is a substrate of the ABC-superfamily xenobiotic transporters ABCB1 (MDR1, Pg-P) and ABCG2 (BCRP). Pregnane X receptor (PXR, NR1I2) and constitutive androstane receptor (CAR, NR1I3) are involved in the control of expression of ABCB1 and ABCG2.
Aim of the study
In this study, we assessed the impact of inherited variants in ABCB1, ABCG2, PXR, and CAR genes on dasatinib efficacy and toxicity in CML.
Materials and methods
Sixty-one tagging SNPs in ABCB1, ABCG2, PXR, and CAR genes were analyzed by real-time quantitative PCR with specific probes in 86 CML patients who failed imatinib therapy.
Results
We found the associations between SNPs rs7787082 (ABCB1, OR = 0.2; 95% CI = 0.06-0.66, p = 0.008), rs12505410 (ABCG2, OR = 3.82; 95% CI = 1.38-10.55; p = 0.010), and rs3114018 (ABCG2, OR = 0.24; 95% CI = 0.08-0.71; p = 0.010) and the probability of achieving CCyR. Furthermore, progression-free survival (PFS) was significantly influenced by SNPs rs3732357 (HR = 0.2, 95% CI = 0.26-0.70; p = 0.001), rs3732360 (HR = 0.59; 95% CI = 0.38-0.93; p = 0.020), rs11917714 (HR = 0.58; 95% CI = 0.36-0.92; p = 0.020), and rs3732359 (HR = 0.57; 95% CI = 0.36-0.91; p = 0.024) in PXR; rs2307418 (HR = 2.02; 95% CI = 1.19-3.43; p = 0.048) in CAR; and rs2235023 (HR = 2.49; 95% CI = 1.13-5.50; p = 0.011) and rs22114102 (HR = 1.90; 95% CI = 1.00-3.63; p = 0.028) in ABCB1. Moreover, overall survival (OS) was impacted by rs3842 (HR = 1.84; 95% CI = 1.01-3.33; p = 0.012) and rs2235023 (HR = 2.28; 95% CI = 1.03 = 5.02; p = 0.027) in ABCB1, rs11265571 (HR = 1.59; 95% CI = 0.82-3.08; p = 0.037) and rs2307418 (HR = 73.68; 95% CI = 4.47-1215.31; p = 0.003) in CAR, and rs3732360 (HR = 0.64; 95% CI = 0.40 = 1.04; p = 0.049) in PXR. Taking into account the influence of the tested SNPs on treatment toxicity, we found a significant relationship between allele G of polymorphism in the ABCB1 rs7787082 (OR = 4.46; 95% CI = 1.38-14.39 p = 0.012) and hematological complications assuming the codominant gene inheritance model as well as a significant correlation between the presence of minor allele (G) of SNP rs2725256 in the ABCG2 gene (OR = 4.71; 95% CI = 1.20-18.47; p = 0.026) and the occurrence of non-hematological complications assuming a recessive gene inheritance model.
Conclusion
Our data suggest that inherited variants in the genes encoding for proteins involved in the transport of xenobiotics may modify the toxicity and efficacy of dasatinib therapy in CML patients.
Keywords: Chronic myeloid leukemia, dasatinib, ABCB1, ABCG2, PXR, CAR, single nucleotide polymorphisms
Introduction
Chronic myelogenous leukemia (CML) is a rare hematological malignancy with an yearly incidence of one per 100,000 individuals (1). The molecular background of the disease has been elucidated since the discovery of the Philadelphia (Ph) chromosome, a consequence of the reciprocal translocation between chromosomes 9 and 22 and the resulting BCR-ABL1 fusion gene that encodes for constitutively activated tyrosine kinase (2, 3). Introduction of imatinib, the first-generation BCR-ABL1 tyrosine kinase inhibitor (TKI), has been a great proof of concept of targeted therapy and has dramatically improved CML prognosis (4). However, approximately one-third of patients develop primary or secondary resistance to imatinib (5). Second-generation TKIs such as dasatinib, and later nilotinib and bosutinib, were introduced to treat imatinib-resistant or intolerant CML patients and subsequently also become first-line treatment options (6, 7).
Dasatinib has a different chemical structure than imatinib and higher potential to inhibit BCR-ABL1 kinase (8). The drug targets a broader spectrum of kinases including SRC kinases, and this contributes to its efficacy as well as specific side effects in CML (9). The pharmacokinetic properties of dasatinib are similar to other TKIs. After oral administration, the molecule binds plasma proteins and is metabolized by liver P450 cytochrome family enzymes (mainly CYP3A4) as well as FMO-3 and UDT-glucuronylotransferase. The process of dasatinib excretion from cells is mediated by ATP-binding cassette (ABC) transporters ABCB1 and ABCG2 (10). ABC transporters are a conserved family of membrane proteins responsible for cell protection against xenobiotics (11).
The expression of genes involved in metabolism and excretion of xenobiotics including the ABC genes is regulated by specific xenosensors—genes that are activated as a response to higher concentrations of xenobiotics. Among them, PXR (NR1I2) and CAR (NR1I3) possess specific DNA-binding domains built up from zinc fingers which enable them to recognize DNA elements, characteristic for enzymes taking part in metabolism of xenobiotic substances (12, 13). PXR and CAR stimulate the expression not only of ABCB1 and ABCG2 but also of cytochrome P450 enzymes and many other genes (14, 15). It was shown that single-nucleotide polymorphisms (SNPs) in the aforementioned genes impact on the metabolism of various drugs (16, 17).
The present study aimed to define genetic markers influencing the outcome of dasatinib therapy in patients with imatinib-resistant or intolerant CML. To that end, we analyzed 61 tagging SNPs in ABCB1, ABCG2, PXR, and CAR genes and studied their effects regarding different parameters of response depth and duration as well as toxicity in CML patients.
Materials and methods
Patients
The study included 86 Polish Caucasian CML patients treated in five tertiary hematological centers in Poland (Department of Hematology, Medical University of Łódź; Department of Hematology, Jagiellonian University Medical College, Kraków; Department of Hematology, Medical University of Gdańsk; Department of Hematology, Copernicus Specialist Municipal Hospital, Toruń; Department of Hematology, Teaching Hospital No 1, Rzeszów). The group included 43 women and 43 men with a median age of 48 years at CML diagnosis (range 18-100). All patients received imatinib at 400 mg/day as a first-line treatment with TKI. The initial dose of dasatinib administered in the second (after imatinib failure) or third (after imatinib and nilotinib failure) line of therapy was 100 mg/day. Complete clinical and laboratory data concerning the course of dasatinib therapy were collected for the analyses of the association with the tested SNPs. Cytogenetic responses were evaluated by classical chromosome banding technique or FISH. The BCR-ABL1 gene expression was assessed by quantitative real-time PCR according to standard protocols described elsewhere (18).
Clinical endpoints
For the purpose of this analysis, we used definitions of treatment endpoints consistent with the European Leukemia Net recommendations (19). Cytogenetic response (CyR) was defined as complete (0% of Ph chromosome, CCyR), partial (1%–34% of Ph chromosome, PCyR), minimal (35%–65% Ph chromosome, mCyR), and no response (>65% of Ph chromosome). Optimal cytogenetic response was categorized as CCyR achievement within 12 months from treatment start. Molecular responses were defined as complete (CMR) when the BCR-ABL1 gene transcript level was below 0.01%, major (MMR) when the BCR-ABL1 gene transcript level was between 0.01% and 0.1%, and no response when the BCR-ABL1 gene transcript level was >0.1%. Optimal molecular response was defined as achievement of at least MMR within 18 months from treatment start. Progression-free survival (PFS) was defined as the interval between start of dasatinib treatment and CML progression or death from any cause, or last follow-up without progression. Overall survival (OS) was counted as the time between dasatinib treatment start and date of death, or last follow-up when the patient was still alive. Analyzed adverse events of the treatment with dasatinib included hematological toxicities of grade 3 or 4 and any non-hematological complication of grades 2–4.
Polymorphism selection
Tagging SNPs for ABCB1, ABCG2, PXR, and CAR were selected using the Tagger algorithm, available through Haploview (20), using pairwise SNP selection with a minimum r2 threshold of 0.8.
The set of common genetic variants (sequences including 5 kb upstream of the first exon and 5 kb downstream of the last exon of each gene), with minor allele frequency (MAF) ≥5% in Caucasians from the International HapMap Project (21), was included for ABCB1, ABCG2, PXR, and CAR. This process resulted in a selection of 26 tagging SNPs for ABCB1 (average r2 of tagging SNPs with the SNPs they tag = 0.958), 17 SNPs for ABCG2 (average r2 = 0.965), 11 SNPs for PXR (average r2 = 0.975), and seven SNPs for CAR (average r2 = 1.000). This selection therefore captures a very high degree of the known common variability in these genes (22).
Genotyping
DNA was isolated from peripheral blood samples using DNA Blood Mini Kit (Qiagen) and genotyped. Duplicates of 8% of the samples were interspersed throughout the plate for ensuring the internal quality controls. Both TaqMan (ABI, Applied Biosystems, Foster City, CA, USA) and KASP (KBioscence, Hoddesdon, UK) technologies were used for genotyping according to the manufacturer’s protocol. PCR plates for TaqMan as well as KASP assays were read on a Viia7 Real-Time PCR platform (Applied Biosystems). The Viia7 RUO Software (Applied Biosystems) was used to determine the genotypes. In our analysis, all individuals with a call rate <80% were excluded from further investigation.
Statistical analysis
Statistical significance of the associations between selected parameters of response to dasatinib therapy and genetic variants was evaluated using R v2.11 software. Logistic regression was used to assess the association between the genetic variability of the SNPs and treatment efficacy, defined by the following endpoints: CCyR at 12 months, MMR at 18 months. Treatment toxicity was investigated with logistic regression as well, using the following endpoints: appearance of any non-hematological toxicities of grades 2–4 including fluid retention, hematological toxicity of grade 3 or 4. These endpoints were used as dichotomous variables. To study the associations between SNPs and PFS and OS, Cox proportional hazard regression was used. SNPs were analyzed according to the following inheritance models: “co-dominant,” where the homozygous major allele (reference category) was compared separately with two different genotypes that include the minor allele (heterozygotes and homozygotes for the variant allele), and “recessive,” whereby the comparison groups were minor homozygous genotypes against the rest (combining heterozygotes and homozygotes for the major allele). The most significant test between the codominant and the recessive genetic models was used to determine the statistical significance of each association of each SNP. All analyses were adjusted by age at diagnosis, sex, CML phase, Sokal score, and use of dasatinib in the second or third line. The results of logistic regression analyses were expressed as odds ratios (OR), and the results of Cox regression analyses were expressed as hazard ratios (HR), with 95% confidence intervals (CI). The results were adjusted for multiple comparisons by Bonferroni correction which was calculated as p = 0.05/(61 SNPs × 2 models) = 0.00041.
Functional characterization of the SNPs
For the SNPs that achieved statistical significance, several online databases were used for analysis of the influence of SNPs on cellular processes such as transcription or association with protein activity.
RegulomeDB is a database that assigns SNPs to predicted and known regulatory sites in the human genome regions that affect transcriptional processes, DNAase hypersensitivity regions, transcription factor binding sites, and promoter regions.
HaploReg is a tool designed to search for significant effects of SNPs in haplotype blocks, including SNPs in disease-related loci, on gene expression levels and protein activity. Using HaploReg, it is possible to assess whether specific SNPs are located in the eQTL (expression quantitative trait loci) loci and to check the effect of SNPs on gene regulatory regions. GTEx is a database that allows to search for relationships between genetic variation (including SNPs) and the expression of genes and proteins in individual tissues.
Results
Clinical data
Eighty-six Polish Caucasian CML patients treated in five tertiary hematological centers in Poland were genotyped within the present study. The characteristics of the study population are described in Table 1 . The majority of patients was diagnosed in the chronic phase (85%). Only 6% of the patients carried additional cytogenetic aberrations; the remainder were characterized only by the presence of the Ph chromosome. Regarding Sokal score, 43% of patients were diagnosed as low risk, and 31% and 26% were characterized as intermediate and high risk, respectively.
Table 1.
Summary of the dasatinib treatment results.
| Dasatinib treatment | Number of patients (%) |
|---|---|
| All | 86 (100%) |
| In the 2nd line | 78 (90.7%) |
| In the 3rd line | 8 (9.3%) |
| Response to treatment a | |
| Optimal CyR b | 52 (60.5%) |
| CCyR | 49 (57%) |
| PCyR | 9 (11%) |
| mCyR | 21 (24%) |
| noCyR | 7 (8%) |
| Optimal MR c | 8 (9.3%) |
| CMR | 24 (30%) |
| MMR | 19 (24%) |
| No MR | 37(46%) |
| Progression | 14 (16.3%) |
| Fluid retention | 15 (17.4%) |
|
Incidence
of any hematological complications |
33 (38.4%) |
Response to treatment: in the database were included results of the last medical visit for each patient.
Optimal CyR: achievement of the complete cytogenetic response within 12 months from treatment start.
Optimal MR: achievement of at least the major molecular response within 18 months from treatment start. Only eight patients (9.3%) achieved optimal response, i.e., at least MR3 within 18 months from the start of treatment; other patients achieved CMR and MMR but after 18 months of treatment, therefore they were not included as optimal MR.
Data on the cytogenetic and molecular response as well as the occurrence of treatment side effects are included.
Eighteen patients (21%) discontinued imatinib due to intolerance to the treatment, 68 patients (79%) due to disease progression. The median duration of imatinib treatment in these patients was 637 days (1.75 years), while the median duration of dasatinib treatment, calculated as the time between the initiation of therapy and the last medical check-up, was 1,492 days (4.09 years). The overall survival and PFS of the cohort is showed in Figure 1 .
Figure 1.
The Kaplan-Meier plots showing overall survival (A) and progression- free survival (B) in the whole cohort of 86 CML patients treated with dasatinib.
All patients received imatinib as first-line therapy of CML in the standard dose of 400 mg once daily and were later treated with dasatinib at 100 mg/day due to the ineffectiveness or intolerance of imatinib. Patients treated with dasatinib right after imatinib treatment (in the second line) accounted for 91% (78 patients) of the group, while eight patients (9%) received dasatinib in the third line, after nilotinib was given for imatinib failure or toxicity.
Sixty-five patients (76%) had no changes in dasatinib dosage while in five cases (6%) the dose was subsequently increased to 140 mg/day at the physician’s discretion and 16 (18%) patients had the dose decreased to 80 mg/day due to toxicity. The main complications that occurred during the follow-up in the present analysis were fluid retention (15 patients, 17%) and hematological complications including thrombocytopenia, cytopenia, neutropenia, lymphopenia, and agranulocytosis (33 patients, 38%). Other undesirable effects included pulmonary hypertension, abdominal pain, arrhythmias, and increased creatine kinase level.
Genotyping quality control
Genotyping of 61 preselected SNPs for ABCB1, ABCG2, CAR, and PXR was successful in all included patients. The average call rate was 98.42% ranging from 85.25% to 100%. Genotype distributions were in accordance with the Hardy–Weinberg equilibrium for all tested loci.
Influence of SNPs on cytogenetic and molecular response to dasatinib
The analysis using a logistic regression model identified the significant impact of tested SNPs on the probability of achieving CCyR following dasatinib treatment. Assuming the codominant model of inheritance, noteworthy associations (p < 0.05) were found between the achievement of CCyR after 12 months of therapy and the following SNPs: ABCB1 gene: rs7787082, PXR rs2461818 and two SNPs in the ABCG2 gene: rs12505410 and rs3109823 ( Table 2 ). Assuming a recessive inheritance model, statistical significance was observed for two SNPs in the ABCG2 gene: rs2622621 and rs3114018 ( Table 2 ). No significant results were found regarding the potential influence of SNPs on the probability of the achievement of MMR after 18 months of dasatinib treatment.
Table 2.
Associations between selected SNPs and cytogenetic response after 12 months of treatment as well as incidence of hematological and non-hematological complications.
| SNP | Gene | Alleles M/m a | Endpoint b | Model c | OR | 95% CI | p-value |
|---|---|---|---|---|---|---|---|
| rs7787082 | ABCB1 | G/A | CyR-12 | CD | 0.20 | 0.06-0.66 | 0.008 |
| rs2461818 | PXR | C/T | CyR-12 | CD | 0.16 | 0.03-0.89 | 0.036 |
| rs12505410 | ABCG2 | T/G | CyR-12 | CD | 3.82 | 1.38-10.55 | 0.010 |
| rs3109823 | ABCG2 | T/C | CyR-12 | CD | 2.87 | 1.11-7.40 | 0.029 |
| rs2622621 | ABCG2 | C/G | CyR-12 | R | 0.21 | 0.05-0.92 | 0.038 |
| rs3114018 | ABCG2 | A/C | CyR-12 | R | 0.24 | 0.08-0.71 | 0.010 |
| rs7787082 | ABCB1 | G/A | HC | CD | 4.46 | 1.38-14.39 | 0.012 |
| rs2725256 | ABCG2 | A/G | NHC | R | 4.71 | 1.20-18.47 | 0.026 |
M, major allele; m, minor allele.
CyR-12, achievement of cytogenetic response within 12 months from treatment start; HC, hematological complications; NHC, non-hematological complications.
CD, codominant; R, recessive.
Influence of SNPs on dasatinib therapy side effects
There was no significant association between tested clinical factors and the occurrence of analyzed side effects including grade 2–4 non-hematological complications or grade 3 and 4 neutropenia or thrombocytopenia. In contrast, a noteworthy correlation was observed between rs7787082, in the ABCB1, and the occurrence of grade 3 or 4 hematological complications assuming the codominant inheritance model. Furthermore, a significant association was found between rs2725256 in ABCG2 and occurrence of non-hematological complications assuming a recessive model of inheritance ( Table 2 ).
Influence of SNPs on OS and PFS on dasatinib therapy
We found a statistically significant influence of Sokal score on OS (HR = 1.34, 95% CI = 1.21-2.09, p = 0.001). No other clinical pretreatment parameter was related to patients’ survival functions. Interestingly, assuming the codominant inheritance model, there was a significant correlation between four SNPs (ABCB1 rs3842, ABCB1 rs2235023, CAR rs11265571, PXR rs3732360) and OS in the patients treated with dasatinib. Furthermore, rs2307418 in the CAR gene impacted OS assuming a recessive inheritance model.
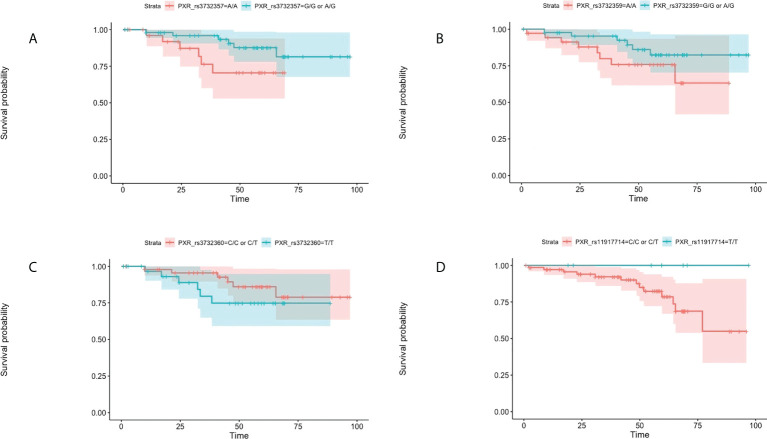
Moreover, seven tested SNPs significantly influenced the probability of PFS. Assuming the codominant model of inheritance, these were the following SNPs: CAR rs2307418 PXR rs3732357, ABCB1 rs2235023 ABCB1 rs22114102 PXR rs3732360, and PXR rs11917714 PXR rs3732359 ( Table 3 and Figure 2 ).
Table 3.
Associations between selected SNPs, overall survival, and progression-free survival during dasatinib treatment.
| SNP | Gene | Alleles M/m a | Endpoint b | Model c | HR | 95%CI | p value |
|---|---|---|---|---|---|---|---|
| rs3842 | ABCB1 | T/C | OS | CD | 1.84 | 1.01-3.33 | 0.012 |
| rs2235023 | ABCB1 | C/T | OS | CD | 2.28 | 1.03-5.02 | 0.027 |
| rs11265571 | CAR | A/T | OS | CD | 1.59 | 0.82-3.08 | 0.037 |
| rs3732360 | PXR | T/C | OS | CD | 0.64 | 0.40-1.04 | 0.049 |
| rs2307418 | CAR | T/G | OS | R | 73.68 | 4.47-1215.31 | 0.003 |
| rs2307418 | CAR | T/G | PFS | CD | 2.02 | 1.19-3.43 | 0.048 |
| rs3732357 | PXR | A/G | PFS | CD | 0.42 | 0.26-0.70 | 0.001 |
| rs2235023 | ABCB1 | C/T | PFS | CD | 2.49 | 1.13-5.50 | 0.011 |
| rs22114102 | ABCB1 | C/T | PFS | CD | 1.90 | 1.00-3.63 | 0.028 |
| rs3732360 | PXR | T/C | PFS | CD | 0.59 | 0.38-0.93 | 0.020 |
| rs11917714 | PXR | C/T | PFS | CD | 0.58 | 0.36-0.92 | 0.020 |
| rs3732359 | PXR | A/G | PFS | CD | 0.57 | 0.36-0.91 | 0.024 |
M, major allele; m, minor allele.
OS, overall survival; PFS, progression-free survival.
CD, codominant; R, recessive.
Figure 2.
Kaplan-Meier plots for progression- free survival in regard to the presence of rs3732357 (A), rs3732359 (B), rs3732360 (C), and rs11917714 (D) in the PXR gene in the co-dominant inheritance model.
Functional SNP annotation
Table 4 shows a summary of the potential functional impact of the tested SNPs based on HaploReg, RegulomeDB, and GTEx databases. The table includes SNPs that showed statistical importance in this analysis.
Table 4.
Summary of bioinformatic SNP annotations.
| SNP ID | Gene | Alleles M/m a | MAF b | Rank c | HaploReg d | eQTL e | GTEx f |
|---|---|---|---|---|---|---|---|
| rs7787082 | ABCB1 | G/A | 0.18 | 6 | CRX FOXD3 GFI1 | – | Testes, skin |
| rs3842 | ABCB1 | T/C | 0.14 | n/a | PLZF | ABCB4 | Brain, nerves |
| rs2235023 | ABCB1 | C/T | 0.09 | 6 | CEBPB ISL2 POU1F1 RHOX11 | ABCB1 ABCB4 | Testes, muscles |
| rs2214102 | ABCB1 | C/T | 0.09 | 4 | GR | – | Heart, colon |
| rs2725256 | ABCG2 | A/G | 0.33 | 6 |
FOXA HNF4 HBP1
POU1F1 RXRA STAT |
SPP1 | Adipocytes |
| rs12505410 | ABCG2 | T/G | 0.40 | 4 | PBX3 | – | Testes, blood |
| rs2622621 | ABCG2 | C/G | 0.30 | 5 | - | – | Blood |
| rs3114018 | ABCG2 | A/C | 0.49 | 5 | LHX3 MEF2 NANOG POU2F2 | – | Esophagus, heart |
| rs3109823 | ABCG2 | T/C | 0.26 | 6 | CTCF NRF-2 YY1 | – | Blood |
| rs2307418 | CAR | T/G | 0.15 | 5 | ERalpha-A GCMGR | TOMM40L USF1 | Brain, muscles, skin |
| rs11265571 | CAR | A/T | 0.17 | 4 | ERalpha-A PAX-5 RHOX11 | Skin, testes, colon | |
| rs2461818 | PXR | C/T | 0.08 | 6 | ARID5B FOXO2 FOXO3 FOXP1 HDAC2 IK2 PLZF SOX6 | PLA1A GSK3B | Thyroid |
| rs11917714 | PXR | C/T | 0.17 | 6 | HIC1 | GSK3B | Nerves, small intestine, esophagus, testes |
| rs3732357 | PXR | A/G | 0.35 | 5 | GLI GLIS2 ZIC | GSK3B | Adipocytes, colon, esophagus, stomach, arteries, thyroid, brain, muscles, lungs |
| rs3732360 | PXR | T/C | 0.23 | 6 | E2A LMO2 MYF TATA | GSK3B GPR156 | Adipocytes, nerves, esophagus, brain, colon, arteries |
| rs3732359 | PXR | A/G | 0.20 | 6 |
BCL NRSF PLAG1
SIN3AK20 TAL1 YY1 |
GSK3B GPR156 | Adipocytes, nerves, esophagus, brain, colon, arteries, thyroid |
The table contains associations for all SNPs, while those that occur only in blood cells are bold.
M: major allele; m: minor allele.
MAF: minor allele frequency in the 1000 Genomes European population.
Rank from RegulomeDB: 1 is given to SNPs showing the strongest evidence of a role in regulating the transcription process by binding transcription factors, while 6 to SNPs with a low probability of influencing to transcription.
HaploReg: the tested SNP probably influences the expression of mentioned genes.
eQTL: the tested SNP is located in the eQTL (expression quantitative trait loci) of mentioned genes.
GTEx: the relationship between the tested SNP and the tissue in which the gene is expressed.
The RegulomeDB portal achieving scores 1–3 indicates the probability of the analyzed SNP belonging to the sequence affecting the binding process of transcription factors. All tested SNPs scored 4, 5, and 6, which means that the binding of transcription factors is very unlikely.
Analysis with HaploReg showed the probable influence of SNPs on the regulation of genes. Noteworthy is the influence of some studied SNPs on the family of FOX transcription factors, which affect a number of cellular processes. All details are included in Table 4 .
In the present work using the GTEx portal, a significant link with the gene expression level was demonstrated for most of the candidate SNPs tested. However, only for SNPs rs12505410, rs2622621, rs3109823 in ABCG2 were the associations in blood cells, while the rest were in tissues not related to CML pathogenesis.
Discussion
We used a candidate gene approach to evaluate the impact of inherited genetic differences on the outcome of CML treatment with dasatinib. The ABCB1 and ABCG2 genes, encoding for known transporters of dasatinib, and PXR and CAR xenosensor genes, with the role of transcription factors for many genes which take part in the pharmacokinetic processes, were chosen for analysis.
A tagging approach was applied to capture the common genetic variability of ABCB1, ABCG2, PXR, and CAR and resulted in a selection of a total of 61 tagging SNPs that were subsequently genotyped in 86 CML patients treated with dasatinib in the second and third lines of treatment.
Significant amount of data has been reported regarding the influence of SNPs in drug transport and metabolism genes on imatinib, another TKI. Kim et al. investigated the influence of SNPs in genes potentially involved in metabolism of imatinib. They found that the rs2231137 GG homozygotes (ABCG2), rs776746 AA (CYP3A5) homozygotes, and advanced stage strongly correlated with poor response to treatment with imatinib; however, the SLC22A1-rs683369 GG homozygotes and advanced stage were correlated with therapy failure (23). Seong et al. analyzed the impact of SNPs in cytochrome P450 enzymes and drug transporters on the imatinib concentration in plasma and clinical response in CML patients. They concluded that rs2231142 (421C>A) situated in ABCG2 is highly associated with MMR achieved by CML patients (24). In contrast, Takahashi et al. showed that homozygotes AA in the same SNP had a higher imatinib concentration than CC (25). In our study, rs2231142 (421C>A) in ABCG2 showed no statistically significant association on dasatinib treatment endpoints.
However, to the best of our knowledge there are little data available regarding influence on inherited background in ABC transporters on dasatinib therapy in CML. Skoglund et al. found that wild-type ABCG2 had a protective effect against the cytotoxicity of all investigated tyrosine kinase inhibitors in exception of bosutinib. Skoglund et al.’s finding of SNPs ABCG2 421C>A, 623T>C, 886G>C, and 1574T>G showed a reduction in ABCG2 cell membrane expression and the protective effect of ABCG2 against imatinib, CGP74588, dasatinib, and nilotinib cytotoxicity (26). In our study, rs2231142 (421C>A) showed no significance regarding dasatinib treatment endpoints.
To our knowledge, there are no publications available on the effect of single-nucleotide polymorphisms in the PXR and CAR nuclear receptor genes on the outcomes of dasatinib treatment in CML patients. However, based on the results of our work, it is assumed that such an influence exists. Analysis of SNPs showed that rs2461818 in PXR has an influence on achieving clinical outcome expressed through CCyR after 12 months (OR = 0.16; 95% CI = 0.03-0.89; p = 0.036). The CAR and PXR proteins belonging to the same family of nuclear receptors are known as transcription factors for genes involved in the metabolism of exogenous substances and their removal from the body (13). As CAR stimulates the expression of proteins related to imatinib metabolism ABCB1, ABCG2, ABCC2, hOCT1, and CYP3A4 (13, 27, 28), genetically influenced changes in the activity of this gene product may have an indirect effect on the plasma concentration of imatinib and thus potentially also the effects of TKI BCR-ABL1 treatment. Although dasatinib belongs to the same group of drugs, its molecular structure and metabolism differ from that of imatinib, which may result in a different response to treatment in the context of the same polymorphic changes, as evidenced by the obtained results.
Loscocco et al., in their study, examined SNPs in genes from the ABC family (ABCB1, ABCG2, ABCC1, ABCC2) to determine their effectiveness in treatment with another tyrosine kinase inhibitor—nilotinib—in a group of 90 CML patients. They found that CC and CT genotypes in ABCC2 rs3740066 as well as the TT genotypes in ABCB1 rs1045642 correlated with a higher probability of achieving MR3 in a shorter time (p = 0.02, p = 0.004, and p = 0.01), where the GG genotype of ABCG2 rs2231137 was associated with a lower probability of MR3 achievement (p = 0.005). Moreover, the ABCC2 rs3740066 CC genotype and the ABCB1 rs1045642 CC and TT genotypes were positively correlated with MR4 achievement (p = 0.02, p = 0.007, and p = 0.003) (29). Our study does not reveal any correlations between examined SNPs and molecular response in patients treated with dasatinib.
Our results suggest that the naturally occurring germline variation in tested genes has influence on such important endpoints of dasatinib therapy in CML as well as probability of cytogenetic and molecular responses, PFS and OS.
To the best of our knowledge, this is the first study on the impact of ABCB1, ABCG2, PXR, and CAR gene polymorphisms on the outcomes of dasatinib treatment in CML. The work clearly shows the influence of SNPs present in genes related to dasatinib metabolism (ABCB1 and ABCG2) and genes encoding transcription factors (PXR, CAR) on treatment outcomes, susceptibility to side effects, and overall survival and progression-free time. Furthermore, the analysis we present here has a number of strengths. First of all, cases were collected in a relatively small number of hematological centers with high medical reference. Information about multiple clinical endpoint variables of patients has been thoroughly compiled in a single database. For each patient, detailed clinical history and treatment history were checked. The collected data showed that the whole group was homogenous, especially in terms of lack of a good response to imatinib. To the best of our knowledge, this is the first study on the impact of ABCB1, ABCG2, PXR, and CAR gene polymorphisms on the outcomes of dasatinib treatment in CML.
The main weakness of this study is the reduced sample size, which limits the statistical power. Taking into account the large number of SNPs included in the study and the different analysis models, the Bonferroni-corrected threshold for statistical significance was rather stringent (p = 0.00041). None of the associations reported here were statistically significant if this threshold is used. However, it has to be kept in mind that CML is not a common disease and is also often treated with imatinib in the first line. Approximately one-third of cases are switched to the next-generation inhibitors (dasatinib, nilotinib, or others) due to lack of response or intolerance to this treatment regimen. We would like to emphasize that the population that participated in this study were patients treated in the first line with imatinib, and in the second (or third) line with dasatinib. Therefore, the above-presented results cannot be directly translated into the population of patients treated in the first line with dasatinib, because the studied population consisted of people who showed intolerance or progression during treatment, which may be of primary origin, regardless of the treatment used.
Another factor that may be a weakness is the fact that only a group of patients of Polish origin was taken into account in the analysis. Therefore, the results cannot be used in comparison to other ethnic groups (e.g., from Asia), whose genetic variability of the tested SNPs and frequency of their occurrence may differ significantly. To confirm this, additional analysis on different populations should be done.
Taking all into account, there is still little known about the impact of inherited changes in genes involved in pharmacokinetic processes in the case of TKI treatment of CML. Our work is one of the first to describe the influence of ABCB1, ABCG2, PXR, and CAR polymorphisms on the effects of dasatinib therapy (expressed through CCyR, PFS, OS) and toxicity of the therapy (expressed by association with hematological and non-hematological complications). Further analyses are needed to confirm these initial results.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
This study was reviewed and approved by Ethical Committee of Medical University of Lodz (RNN/168/13/KE; 18 June 2013) Łódź, gen. Józefa Hallera Square 1B bioetyka@umed.lodz.pl. The patients/participants provided their written informed consent to participate in this study.
Author contributions
KJ conceived and designed the study. AM performed the lab work and prepared the database. FC performed the statistical analysis. AM analyzed the results and drafted the manuscript. FC and KJ reviewed and edited the manuscript. All other authors provided samples and data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants of Young Hematologists Club of Polish Society of Hematology and Transfusiology Medicine.
Acknowledgments
The authors would like to express their deep gratitude to Dr. Alessandro Martino for his involvement in the project of evaluation polymorphisms in chronic myeloid leukemia. The authors would also like to extend their thanks to Angelika Stein, technician at the DKFZ Genomic Epidemiology Group, for her patience and help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER cancer statistics review, 1975-2009 (Vintage 2009 populations). Natl Cancer Inst (2012). [Google Scholar]
- 2. Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst (1960) 25:85–109. [PubMed] [Google Scholar]
- 3. Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and giemsa staining. Nature (1973) 243(5405):290–3. doi: 10.1038/243290a0 [DOI] [PubMed] [Google Scholar]
- 4. Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, et al. Effects of a selective inhibitor of the abl tyrosine kinase on the growth of bcr-abl positive cells. Nat Med (1996) 2(5):561–6. doi: 10.1038/nm0596-561 [DOI] [PubMed] [Google Scholar]
- 5. Hochhaus A, Hughes T. Clinical resistance to imatinib: mechanisms and implications. Hematol Oncol Clin North Am (2004) 18:641–56. doi: 10.1016/j.hoc.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 6. Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, et al. Discovery of n-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino) thiazole-5 carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem (2004) 47(27):6658–61. doi: 10.1021/jm049486a [DOI] [PubMed] [Google Scholar]
- 7. Deremer DL, Ustun C, Natarajan K. Nilotinib: a second-generation tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia. Clin Ther (2008) 30(11):1956–75. doi: 10.1016/j.clinthera.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 8. Muller MC, Cortes J, Kim D-W, Druker BJ, Erben P, Pasquini R, et al. Dasatinib efficacy in patients with chronic myeloid leukemia in chronic phase and preexisting BCR-ABL mutations (abstract 449). Blood (2008) 112(11):171. [Google Scholar]
- 9. Manley PW, Cowan-Jacob SW, Mestan J. Advances in the structural biology, design and clinical development of bcr-abl kinase inhibitors for the treatment of chronic myeloid leukaemia. Biochim Biophys Acta (2005) 1754:3–13. doi: 10.1016/j.bbapap.2005.07.040 [DOI] [PubMed] [Google Scholar]
- 10. Kamath AV, Wang J, Lee FY, Marathe PH. Preclinical pharmacokinetics and in vitro metabolism of dasatinib (BMS-354825): A potent oral multi-targeted kinase inhibitor against SRC and BCR-ABL. Cancer Chemother Pharmacol (2008) 61(3):365–76. doi: 10.1007/s00280-007-0478-8 [DOI] [PubMed] [Google Scholar]
- 11. Sharom FJ. ABC Multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics (2008) 9(1):105–27. doi: 10.2217/14622416.9.1.105 [DOI] [PubMed] [Google Scholar]
- 12. Liddle C, Goodwin B. Regulation of hepatic drug metabolism: role of the nuclear receptors PXR and CAR. Semin Liver Dis (2002) 22(2):115–22. doi: 10.1055/s-2002-30098 [DOI] [PubMed] [Google Scholar]
- 13. Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA, et al. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol (2002) 62(3):638–46. doi: 10.1124/mol.62.3.638 [DOI] [PubMed] [Google Scholar]
- 14. Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem (2001) 276(18):14581–7. doi: 10.1074/jbc.M010173200 [DOI] [PubMed] [Google Scholar]
- 15. Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med (2001) 7(5):584–90. doi: 10.1038/87912 [DOI] [PubMed] [Google Scholar]
- 16. Zazuli Z, Barliana MI, Mulyani UA, Perwitasari DA, Ng H, Abdulah R, et al. Polymorphism of PXR gene associated with the increased risk of drug-induced liver injury in Indonesian pulmonary tuberculosis patients. J Clin Pharm Ther (2015) 40(6):680–4. doi: 10.1111/jcpt.12325 [DOI] [PubMed] [Google Scholar]
- 17. Moon JY, Chang BC, Lee KE, Bang JS, Gwak HS. Effects of pregnane X receptor genetic polymorphisms on stable warfarin doses. J Cardiovasc Pharmacol Ther (2015) 20(6):532–8. doi: 10.1177/1074248415578906 [DOI] [PubMed] [Google Scholar]
- 18. Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of 'real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe against cancer program. Leukemia (2003) 17(12):2318–57. doi: 10.1038/sj.leu.2403135 [DOI] [PubMed] [Google Scholar]
- 19. Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia (2020) 34(4):966–84. doi: 10.1038/s41375-020-0776-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Available at: http://www.broad.mit.edu/mpg/tagger/ http://www.broad.mit.edu/mpg/haploview/.
- 21.Available at: https://pubmed.ncbi.nlm.nih.gov/14685227/.
- 22. Campa D, Butterbach K, Slager SL, Skibola CF, de Sanjosé S, Benavente Y, et al. A comprehensive study of polymorphisms in ABCB1, ABCC2 and ABCG2 and lung cancer chemotherapy response and prognosis. Int J Cancer (2012) 131(12):2920–8. doi: 10.1002/ijc.27567 [DOI] [PubMed] [Google Scholar]
- 23. Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res (2009) 15(14):4750–8. doi: 10.1158/1078-0432.CCR-09-0145 [DOI] [PubMed] [Google Scholar]
- 24. Seong SJ, Lim M, Sohn SK, Moon JH, Oh SJ, Kim BS, et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol (2013) 24(3):756–60. doi: 10.1093/annonc/mds532 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi N, Miura M, Scott SA, Kagaya H, Kameoka Y, Tagawa H, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet (2010) 55(11):731–7. doi: 10.1038/jhg.2010.98 [DOI] [PubMed] [Google Scholar]
- 26. Skoglund K, Boiso Moreno S, Jönsson JI, Vikingsson S, Carlsson B, Gréen H, et al. Single-nucleotide polymorphisms of ABCG2 increase the efficacy of tyrosine kinase inhibitors in the K562 chronic myeloid leukemia cell line. Pharmacogenet Genomics (2014) 24(1):52–61. doi: 10.1097/FPC.0000000000000022 [DOI] [PubMed] [Google Scholar]
- 27. Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, et al. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem (2002) 277(4):2908–15. doi: 10.1074/jbc.M109326200 [DOI] [PubMed] [Google Scholar]
- 28. Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol (2001) 41:123–43. doi: 10.1146/annurev.pharmtox.41.1.123 [DOI] [PubMed] [Google Scholar]
- 29. Loscocco F, Visani G, Ruzzo A, Bagaloni I, Fuligni F, Galimberti S, et al. Clinical relevance of ABCB1, ABCG2, and ABCC2 gene polymorphisms in chronic myeloid leukemia patients treated with nilotinib. Front Oncol (2021) 11:672287. doi: 10.3389/fonc.2021.672287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.