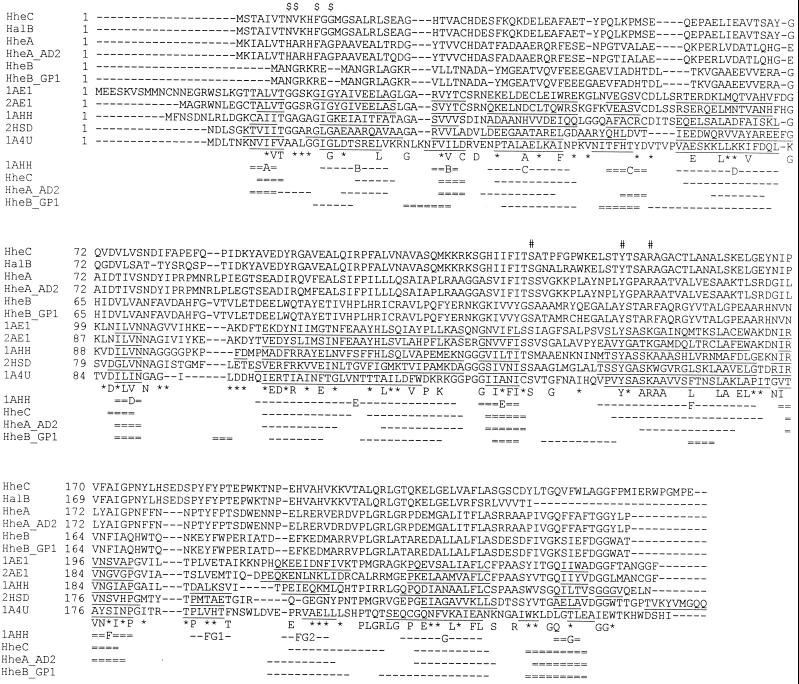
FIG. 1.
Sequence alignment of halohydrin dehalogenases with SDRs with known three-dimensional structures. The sequences were aligned by using the multiple alignment program ClustalW and are shown in decreasing sequence similarity to HheC. Amino acids that are identical in six or more sequences are depicted below the sequence. Positions at which six or more similar residues occur are indicated by an asterisk. The position of the G/A-G/A-X-X-G/A-X-G/A fingerprint typical of the Rossman fold in the SDR family is indicated by $ on top of the alignment. The proposed active site residues in halohydrin dehalogenases are indicated by #. The structural elements identified in three-dimensional structures of SDR family members are underlined. The first line below the sequence alignment shows the secondary structure elements and the nomenclature of 7α-hydroxysteroid dehydrogenase (29). β-Strands are indicated as double broken lines, and α-helices are indicated as single broken lines. The second, third, and fourth lines show the predicted secondary structure elements for HheC, HheAAD2, and HheBGP1, respectively. The nomenclature of the sequences is explained in Table 1.