Figure 2.
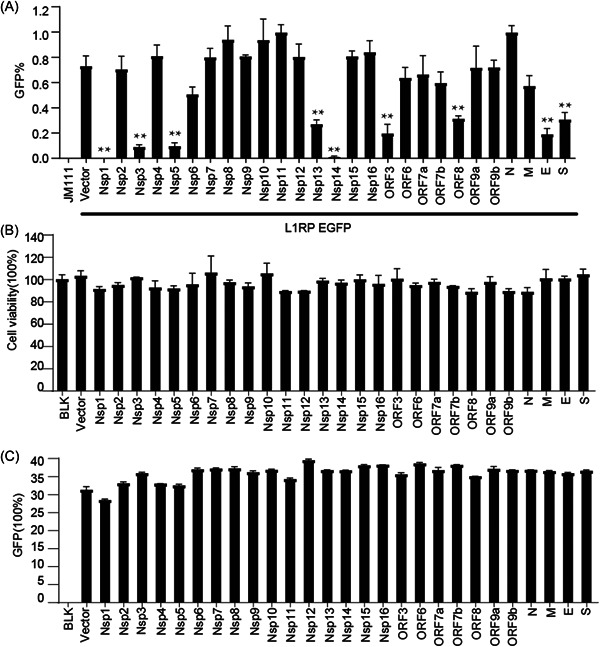
SARS‐CoV‐2‐encoded proteins decrease L1‐retrotransposition frequency. (A) Empty vectors or SARS‐CoV‐2 protein‐expressing plasmids (50 ng) were cotransfected into HEK293T cells along with L1RP EGFP plasmids (1 µg), and the number of EGFP‐positive cells was determined at 4 days posttransfection by flow cytometry. JM111 was used as a negative control for flow cytometric gating. The bar represents retrotransposition efficiency. Experiments were performed in triplicate, and each error bar indicates the standard deviation of three replicates for one experiment. **p < 0.01, Student's t test. (B) HEK293T cells pretransfected with empty vectors or SARS‐CoV‐2 protein‐expressing constructs were seeded on 96‐well plates and cultured for 4 days, followed by MTS staining and measurement of absorbance. The bar represents cell viability. The control‐treated sample was set to 100%. (C) pcDNA3.1‐EGFP plasmids and empty vectors/SARS‐CoV‐2 protein‐expressing plasmids were cotransfected into HEK293T cells, and the number of EGFP‐positive cells was determined at 4 days posttransfection using flow cytometry. The bar represents the percentage of GFP‐positive cells. EGFP, enhanced green fluorescent protein; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; MTS, 3‐(4,5‐dimethylthiazol‐2‐yl)−5‐(3‐carboxymethoxyphenyl)−2‐(4‐sulphophenyl)−2H‐tetrazolium; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
