Figure 4.
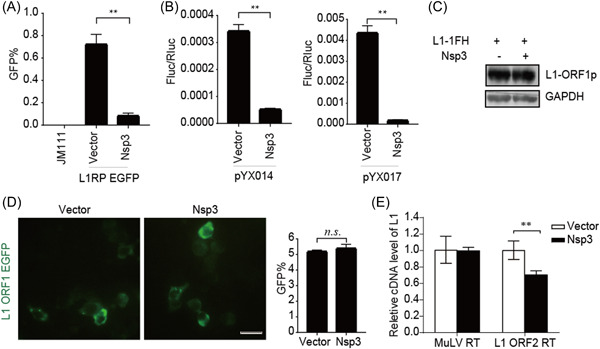
SARS‐CoV‐2 Nsp3 restricts L1 mobility in an ORF2p‐dependent manner. (A) L1RP EGFP plasmids and empty vectors/SARS‐CoV‐2 Nsp3‐expressing constructs were transfected into HEK293T cells, with JM111 used as a negative control, and flow cytometry was performed at 4 days posttransfection. The bar graph depicts the percentage of GFP‐positive cells. **p < 0.01. (B) Empty vector or SARS‐CoV‐2 Nsp3‐expressing constructs were cotransfected along with the pYX014/pYX017 reporter constructs into HEK293T cells, followed by dual‐luciferase assays performed at 4 days posttransfection. The bar graph represents the retrotransposition frequency. **p < 0.01. (C) HEK293T cells were cotransfected with empty vectors/SARS‐CoV‐2 Nsp3 and pc‐L1‐1FH, after which cells were collected at 48‐h posttransfection, and anti‐HA antibodies were used in an immunoblot assay. (D) HEK293T cells were transfected with pEGFP‐N1‐ORF1‐EGFP and empty vectors/SARS‐CoV‐2 Nsp3‐expressing constructs, followed by living‐cell imaging performed at 48‐h posttransfection (scale bar, 20 mm). The bar graph depicts the percentage of GFP‐positive cells according to flow cytometry. (E) LEAP assays, followed by analysis of MuLV RTase and LEAP products by qRT‐PCR. The bar graph of the relative cDNA level of L1 represents reverse‐transcription efficiency of the L1 ORF2p or MuLV. The relative cDNA level of MuLV reverse transcription was set to 1.0. **p < 0.01. cDNA, complementary DNA; EGFP, enhanced green fluorescent protein; LEAP, L1 element amplification protocol; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; Nsp, nonstructural protein; ORF, open reading frame; n.s., not significant; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
