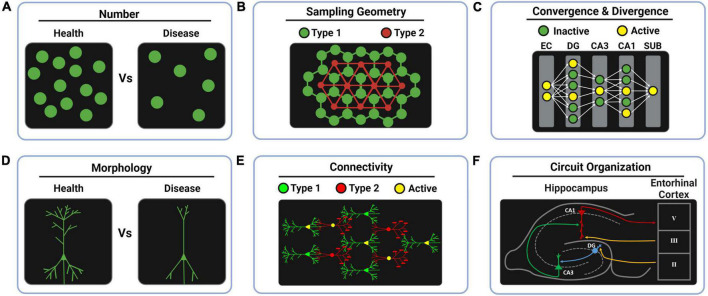
FIGURE 1.
Whole-brain anatomical features for computational neuroscience. (A) The computational redundancy of the brain can be studied by cell counting in health and disease to predict the minimal percentage of neuronal loss that leads to symptoms. (B) Studying the sampling patterns that the brain uses for covering the space can reveal different design principles across brain regions such as the regular mosaic principle found in tagged functional cell types of the retina. In the drawings, lines between cells represent spatial distances, not real connections. (C) Encoding transformations can be interrogated by studying increments (divergence) or decrements (convergence) in neuron number across layers. Likewise, sparse coding and dense coding strategies might be revealed by tagging the percentage of active neurons in each layer during behavioral tasks. (D) The computational impact of morphological features can be studied by comparing health and disease models. (E) Data-driven morpho-neuronal models can be elaborated to understand the design principles behind structural connectivity. (F) Genetically labeling functional cell types provides a way to ultimately study the fine circuit organization of complex brain regions. EC, entorhinal cortex; DG, dentate gyrus; CA3, Ammon’s horn field 3; CA1, Ammon’s horn field 1; SUB, subiculum.