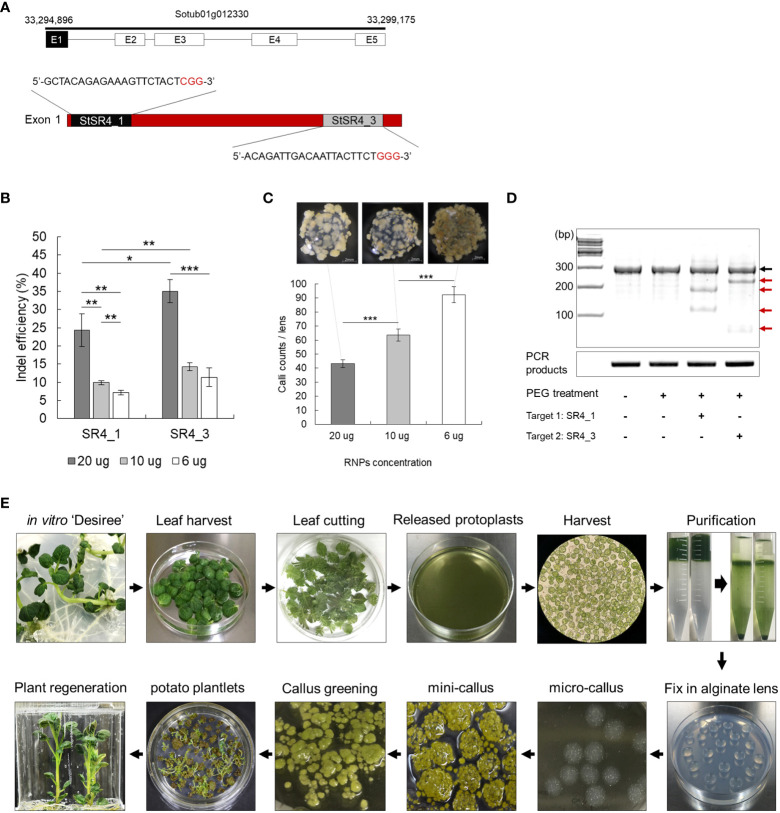
Figure 1.
Ribonucleoprotein (RNP)-mediated CRISPR/Cas9 genome editing in potato. (A) Identification of target susceptibility (S) genes. Selection and design of two crRNA target sites in exon 1 of StSR4 in the potato cultivar Desiree. Black sequence, crRNA target sequence; red sequence, PAM site. (B) Indel efficiency at two target sites, depending on the concentration of the Cas9 protein and guide RNA (gRNA) in protoplasts. Values represent the mean ± SE of three replicated experiments. Student’s t-test; *p < 0.05, **p < 0.01, ***p < 0.001. (C) Efficiency of mini callus induction, depending on the concentration of RNPs in protoplasts. Callus growth as a function of RNP concentration: 20 µg, 10 µg, and 6 µg. (D) The T7E1 assay of mutations in protoplasts independently treated with SR4_1 and SR4_3 RNPs. Black arrow indicates the wild-type fragment; red arrow indicates indel-carrying fragments. (E) Schematic diagram showing the process of inducing plant regeneration from potato protoplast.