Figure 1.
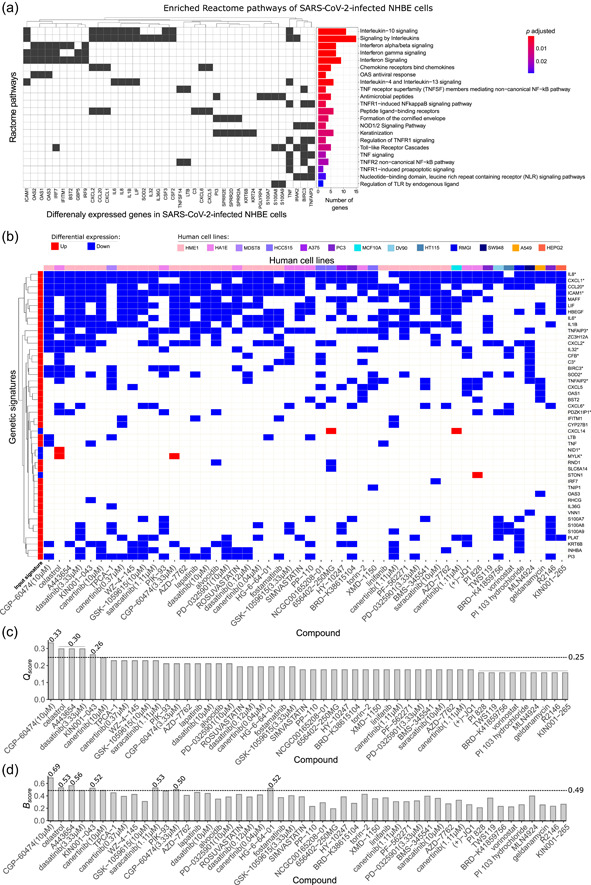
Assessment of gene signature in SARS‐CoV‐2‐infected human bronchial epithelial cells (NHBE) for drug discovery to treat COVID‐19. (a) Enriched biological pathways associated with COVID‐19 from Reactome annotations (p adj < .05) from DEGs identified in NHBE‐infected cells transcriptome data. The genes that enriched each pathway (left) are indicated together with statistical results and pathway description (right). (b) Heatmap of genes from the 50 best‐ranked compound signatures that reversed the genetic signature of SARS‐CoV‐2‐infected NHBE cells in decreasing order of Q score. The signature map annotations are related to up‐ and downregulated genes, and cell lines are indicated in different colors. (c) Compounds in decreasing order of Q score following the output of L1000CDS.2 A dashed line indicates the mean Q score (0.26) threshold. Equal Q score values are displayed over the bars. (d) B score for each compound, considering the enrichment analysis and reverse signature. The dashed line corresponds to the mean B score (0.49). *Genetic signature that justified the biological validation of celastrol. COVID‐19, coronavirus disease 2019; DEGs, differentially expressed genes; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
