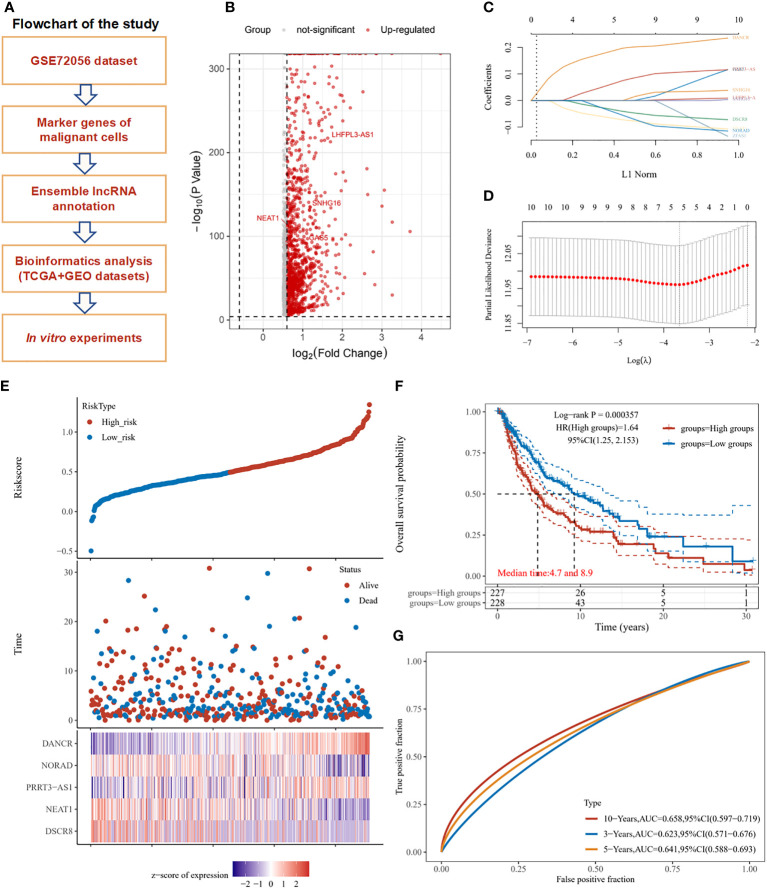
Figure 1.
Study workflow and marker lncRNAs prognosis model construction in SKCM. (A) Workflow of the study. (B) Volcano plot exhibits the marker RNAs of melanoma cells in GSE72056. (C) LASSO coefficient profiles of the marker lncRNAs for overall survival in GSE72056. (D) Partial likelihood deviance of overall survival for the LASSO coefficient profiles. Five features with non-zero coefficients were selected by optimal lambda. (E) Melanoma patients were sorted by risk score, based on which patient status and the selected lncRNA expression of each patient was exhibited. Distribution of risk score (upper figure), OS (middle figure), and heat map of DANCR, NORAD, PRRT3-AS1, NEAT1, and DSCR8 expression in the GSE72056 dataset (lower figure). (F) Survival curves and patient status of 5-marker-lncRNAs. (G) ROC curves compare the prognostic accuracy of the classifier in melanoma patients using AUCs at 1, 5, and 10 years to assess prognostic accuracy.