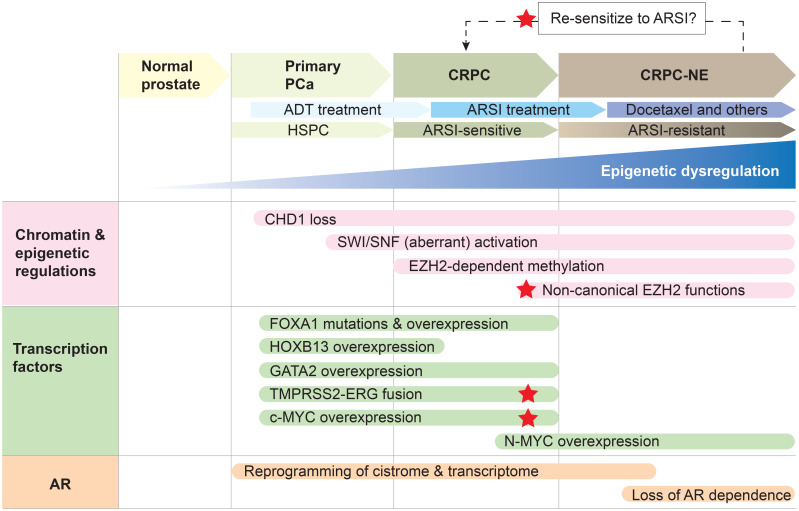
Figure 2.
Epigenetic dysregulation and concordant aberrant transcription factor activity result in a reprogrammed AR cistrome and transcriptome to drive prostate cancer progression. Primary prostate cancer (PCa) is hormone-sensitive (HSPC) and responds to androgen-deprivation therapy (ADT). Resistance to ADT results in the development of castration-resistant prostate cancer (CRPC), which remains reliant on AR signaling and is therefore responsive to second-generation AR signaling inhibitors (ARSI; e.g., abiraterone acetate, enzalutamide). However, resistance to ARSI is accompanied by the onset of CRPC with neuroendocrine features (CRPC-NE), which can be treated with chemotherapy such as docetaxel. Reprogramming the cancer cell to an ARSI-sensitive state is an attractive therapeutic strategy, and red stars denote key players that may be targetable to achieve a reversal in treatment-resistance. Note that this is one example of how prostate cancer may be treated, with an emphasis on how the AR signaling axis is therapeutically targeted. Epigenetic dysregulation underlies prostate cancer progression and the development of treatment resistance to ADT and second-generation ARSI. CHD1 loss, accompanied by the overexpression and aberrant activities of the SWI/SNF remodeling complexes, EZH2 and PRC2, result in a plastic epigenome. Epigenomic plasticity also provides cancer cells the opportunity to develop AR-independent mechanisms of tumor growth when the AR signaling pathway is exposed to more extensive inhibition through ARSI treatment. This, in combination with the hyper-activity or altered activity of pioneer transcription factors FOXA1 and HOXB13 as well as the transcription factors ERG, c-MYC and N-Myc, result in enhancer rewiring and reprogramming of the AR cistrome, thereby driving disease progression and unfavorable therapeutic responses. The pioneer transcription factor GATA2 is a key cofactor for maintaining the AR transcriptional program during HSPC and CRPC — however, it does not seem to reprogram the AR cistrome in the same way that FOXA1 and HOXB13 do, suggesting there may be a hierarchy in pioneer transcription factor activity.