FIGURE 1.
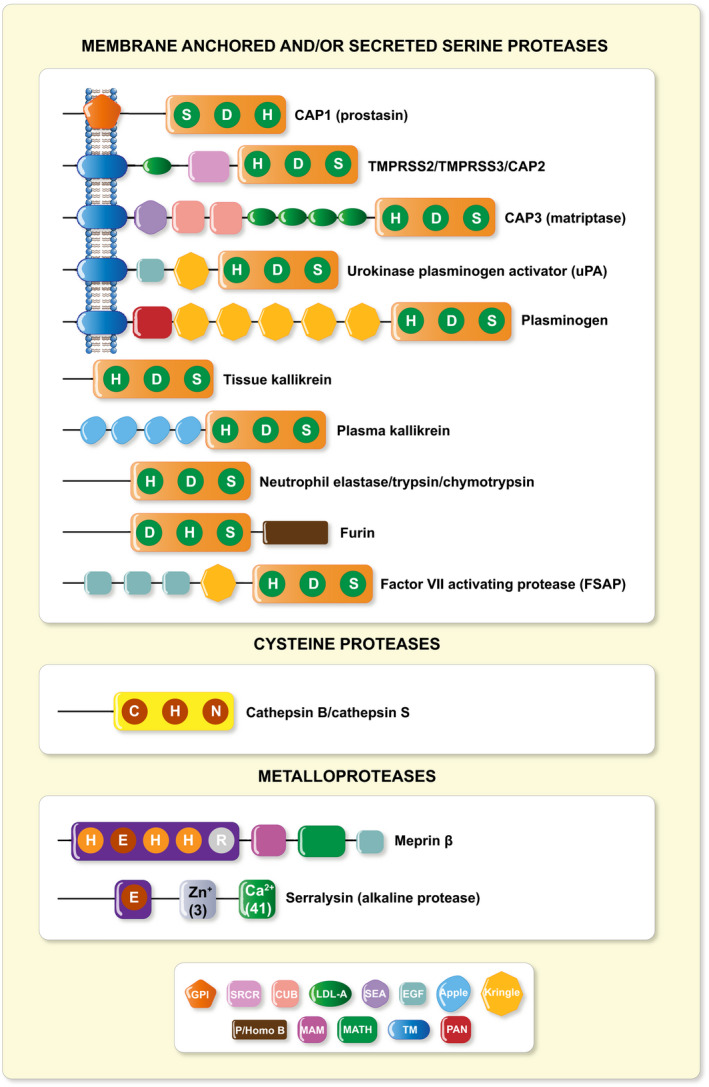
Structural schematic of identified ENaC CAPs. CAP1 (Prss8, prostasin), 4 , 5 CAP2 (Tmprss4), 6 CAP3 (St14/Matriptase), 6 Tmprss3, 7 Tmprss2, 8 , 9 uPA (urokinase‐type plasminogen activator), 10 , 11 plasminogen, 12 , 13 , 14 , 15 trypsin, 16 chymotrypsin, 14 tissue 17 , 18 and plasma kallikrein, 19 elastase, 20 furin, 21 factor VII activating protease, 22 cathepsin B, 23 , 24 cathepsin S, 14 meprin β 25 and serralysin. 26 Predicted structural domains of mouse proteases are indicated 27 : CUB, complement C1r/C2s, urchin embryonic growth factor, bone morphogenic protein 1; EGF, epidermal growth factor‐like; apple; kringle; GPI, glycophosphatidylinositol anchor; LDL‐A, low density lipoprotein A; MAM, meprin, A5 protein, receptor protein phosphatase μ; MATH, meprin and TRAF‐C homology; P/Homo B, paired basic amino acid residue‐cleaving enzyme/homo sapiens B; PAN, PAN/apple; SEA, sperm protein, enterokinase and agrin; SRCR, scavenger receptor cysteine‐rich; TM, transmembrane
