FIGURE 4.
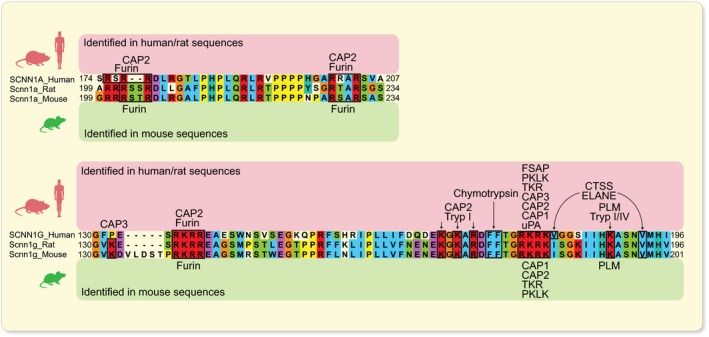
In vitro identified consensus sites for proteolytic ENaC cleavage by CAPs. Alignment of human, rat and mouse α‐ and γ‐ENaC subunits showing a high degree of conservation between species at cleavage sites. Amino acid numbering at the end of transcripts indicates the portion of sequence analysed. CAPs shown to cleave human/rat (indicated above) and mouse (indicated below) ENaC consensus sites in vitro are specified. 3 , 37 , 38 , 39 CTSS, cathepsin S; ELANE, neutrophil elastase; FSAP, factor VII‐activating protease; PKLK, plasma kallikrein; PLM, plasmin; TKR, tissue kallikrein; Tryp, trypsin; uPA, urokinase‐type plasminogen
