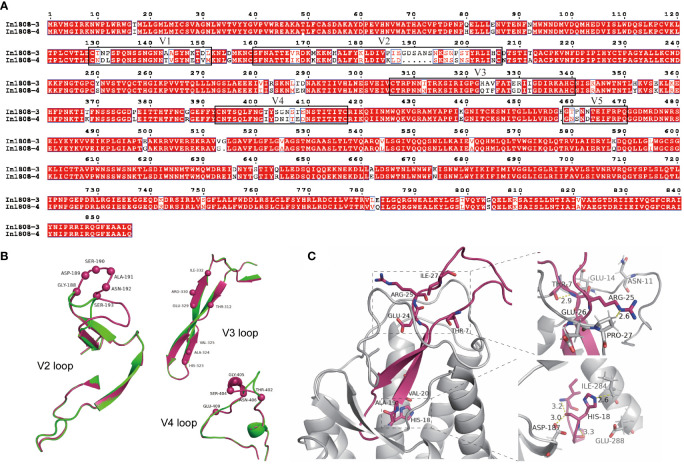
Figure 4.
Sequence and structural analyses of R5 and X4 Envs from subject 1808. (A) Full-length Env amino acid alignment of pseudoviruses In1808-3 (X4) and In1808-4 (R5). Variable regions V1–V5 are shown in boxes. (B) The structural alignment of V2 loop, V3 loop, and V4 loop; the whole structure is shown in cartoon, the main difference amino acids are highlighted by spheres model, and In1808-3 in warm pink, In1808-4 in green. (C) Structural analysis for the V3 region features points in binding of co-receptors CXCR4 using the V3-docking model. The whole structure is shown in cartoon, and the key residues are shown in sticks.