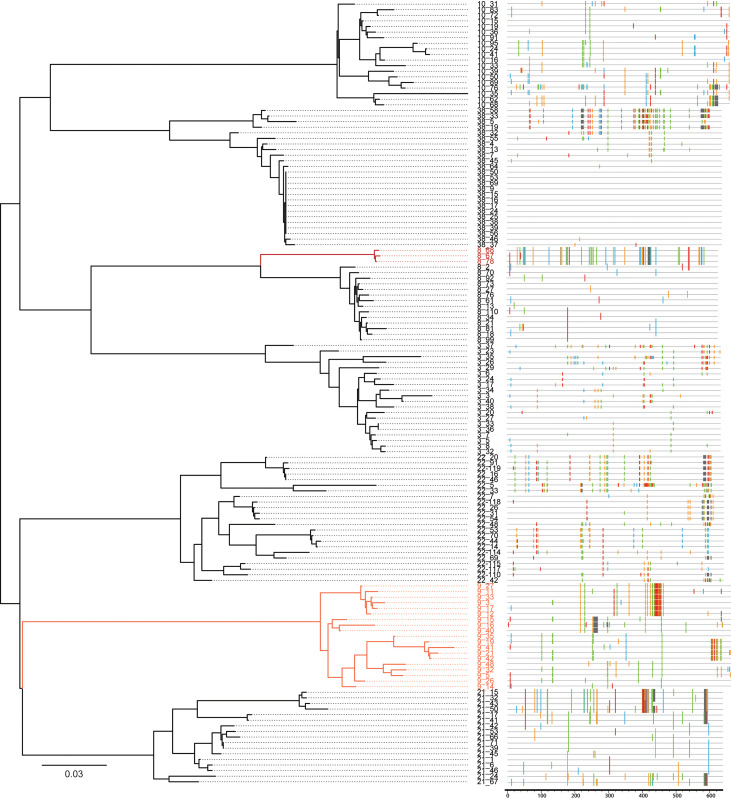
Figure 5.
SGA to detect the HIV-1 variant populations with different coreceptor usages. SGA-derived env sequences from seven HIV-1-infected plasma samples were examined by phylogenetic tree construction (left panels) and Highlighter plot analysis (right panels). Sequences predicted to be X4-tropic are in red face, and the darker the color, the lower the FPR (0.8% and 1.7%). The corresponding Highlighter diagrams denote the locations of nucleotide sequence substitutions in each env sequence in comparison to their consensus sequence (the consensus sequence for each individual is not shown). Nucleotide substitutions and gaps are color coded (A, green; C, blue; G, orange; T, red; and GAP, gray). In individual 1808, both R5-tropic (FPR = 88.5%) and X4-tropic (FPR = 0.8%, in deep red) strains were present and their sequences were quite different. All the sequences are predicted to be X4-tropic (FPR = 1.7%, in light red) in individual 1809.