Abstract
Here, we present a longitudinal shotgun sequencing metagenomics study of 16 healthy, Danish women in the reproductive age. The aim of the study was to investigate whether lactobacilli, orally consumed, had any impact on the vaginal microbiome and its functional potential. The 16 women aged 19–45 years were recruited from Copenhagen, Denmark. One baseline vaginal sample (Day 0) and two study samples (Days 25–30 and Days 55–60, respectively), were sampled. The vaginal samples were analyzed by shotgun metagenomics. We detected 26 species in the vaginal microbiota of the 16 women, of which six belonged to the Lactobacillus genus. We observed three vaginal microbiome clusters mainly dominated by Gardnerella vaginalis, Lactobacillus iners, or Lactobacillus crispatus. The oral probiotic had no detectable effect on either the composition or the functional potential of the vaginal microbiota. Most of the study subjects (11 out of 16 women) exhibited only minor changes in the vaginal microbiome during the treatment with probiotics. Any compositional changes could not be associated to the probiotic treatment. Future studies may benefit from an increased number of participants, and administration of the probiotics during conditions with bacterial imbalance (e.g., during/after antibiotic treatment) or the use of different Lactobacillus spp. known to colonize the vagina.
Keywords: Vaginal microbiome, healthy microbiome, women, public health, shotgun metagenomics
In the vagina, as in other parts of the body, microbes exist in a delicate mutualistic relationship which represents the first line of defense against colonization and infection by opportunistic pathogens, a phenomenon known as colonization resistance [1, 2, 3]. In addition, Lactobacillus spp. (Lactobacillus crispatus, L. gasseri, L. iners or L. jensenii) play an important physiological role in maintaining a low pH (<4.5) in the vagina via production of lactic acid [4, 5, 6]. This production of lactic acid, combined with hydrogen peroxide and bacteriocins, protects the vaginal environment from invasion by other microorganisms, infection of vaginal epithelium and has accordingly been associated with an overall well‐being [2, 4, 5]. Vaginal microbiota not only dominated by a single species but also found with anaerobes and strict anaerobes species (e.g., Gardnerella, Atopobium, Mobiluncus, Prevotella, and Clostridiales spp.), have traditionally been associated with vaginal symptoms consistent with bacterial vaginosis (BV), including discharge, odor, and irritation [2, 3]. Furthermore, women with a vaginal microbiome dominated by G. vaginalis and other anaerobic bacteria are considered to be at risk of adverse events. Yet, recent studies have identified Gardnerella spp. as a sub‐dominant species in the vaginal microbiome of healthy women in the reproductive age [7, 8]. Thus, the overall microbiome functional potential is important to attain a healthy vaginal flora. As such, probiotic may impact the vaginal microbiome indirectly. Furthermore, Reid et al. found that the numbers of vaginal lactobacilli increased significantly in healthy women receiving Lactobacillus orally [4, 5, 9]. Only a few studies investigated the vaginal microbiota of healthy women longitudinally, and very few have tested the impact of probiotics on healthy individuals. Furthermore, many of these studies have been carried out using 16S rRNA gene high‐throughput sequencing [6, 7, 8]. Shotgun metagenomic sequencing has become an acknowledged high‐throughput technology for characterizing microbial communities [6].
The purpose of this study was to investigate, if lactobacilli from oral probiotics had any impact on the structure of the vaginal microbiome and its functional potential, using shotgun‐metagenomics data analysis, a method previously validated on complex biological samples [10, 11].
MATERIALS AND METHODS
Participants and study design
Healthy (self‐reported), reproductive‐aged women were recruited from Copenhagen, Denmark, through online and print advertisements. Women were interviewed and screened at Department of Clinical Microbiology at either Herlev Hospital or Rigshospitalet in Copenhagen, Denmark. Further information can be found in Supplementary material.
One baseline vaginal sample (Day 0) and two study samples (Days 25–30 and Days 55–60, respectively), were sampled from each participant. Participants performed swabs 7–10 days after the final day of menstruation. We used OMNIgene Vaginal kit for the easy self‐collection and stabilization of DNA for microbiome analysis (DNA Genotek, Ottawa, Canada). This allowed for home‐sampling. Instructions for vaginal swabs were given according to the guidelines provided by the manufacturer and is briefly described in Supplementary material. After collection, the samples were stored at −20 °C and mailed (within 7 days) to Department of Clinical Microbiology, Rigshospitalet for storage at −80 °C until all samples had been collected.
Bifodan A/S determined the viability of Lactobacillus spp. in the oral capsules prior to our study (for further details, see Supplementary material).
Furthermore, see Supplementary material for descriptions of molecular microbiology methods.
Statistical analyses
To identify differentially abundant MGSs between the sampling (visits 1, 2, and 3), we performed paired Wilcoxon signed‐rank tests. We specifically tested whether the L. gasseri abundance changed significantly after visit 1. Where multiple hypotheses were evaluated in parallel, the Benjamini–Hochberg method was used to control false discovery rate considering significant all hits below an FDR of 5%.
Ethics
Research ethical approvals were given by the Regional Committee of Danish Data Protection Agency and the Regional Committee of Health Research Ethics for the Capitol Region of Denmark (Trial registration ID No. H‐17004182). Informed consent for participation and publication was obtained from all participants prior to enrollment. Thus, all processes were performed according to the Declaration of Helsinki and its amendments.
RESULTS
Participants
We initially recruited 21 healthy women (age range: 18–45 years), of which 3 dropped out before completing the study and 2 other completed but their swabs were lost during shipment to the department of Clinical Microbiology, Rigshospitalet. Thus, 16 women completed the study (Table 1) (they had not used either any oral or intravaginal antibiotic or antifungal treatments 6 months prior to the study or any oral or intravaginal probiotics 3 months prior to the study).
Table 1.
Study participants overview
| Participant number | Ethnicity | Status | Age (years) | Three vaginal swabs 7–10 days after last day of menstruation | Two rectal swabs performed with 1st. And 3rd. Vaginal swabs |
|---|---|---|---|---|---|
| 1 | Caucasian | Completed | 26 | X | X |
| 2 | Caucasian | Completed | 25 | X | |
| 4 | Caucasian | Completed | 23 | X | |
| 5 | Caucasian | Completed | 36 | X | X |
| 6 | Caucasian | Completed | 24 | X | |
| 8 | Caucasian | Completed | 24 | X | X |
| 9 | Caucasian | Completed | 34 | X | |
| 10 | Caucasian | Completed | 31 | X | X |
| 11 | Caucasian | Completed | 27 | X | |
| 14 | Caucasian | Completed | 32 | X | X |
| 15 | Caucasian | Completed | 37 | X | X |
| 16 | Caucasian | Completed | 34 | X | |
| 17 | Caucasian | Completed | 26 | X | X |
| 18 | Arabic | Completed | 35 | X | X |
| 19 | Caucasian | Completed | 45 | X | X |
| 20 | Caucasian | Completed | 24 | X | |
| 7 | Asian | Lost in mail | 33 | ||
| 13 | Caucasian | Lost in mail | 36 | ||
| 3 | Caucasian | Dropped out | 19 | ||
| 12 | Caucasian | Dropped out | 22 | ||
| 21 | Caucasian | Dropped out | 27 |
Naturally occurring L. rhamnosus was not detected in any of the vaginal samples. However, the species L. gasseri was detected in 33 of the 48 samples (11 out of 16 subjects), ranging from 0.003% to 32.2% relative abundance (Fig. 1). Women with L. gasseri detected in their vaginal microbiota displayed this species already before probiotic consumption (visit 1). We did not observe a significant difference in L. gasseri abundance comparing visit 1 to visit 2 (Wilcoxon signed‐rank test; p = 0.69) or visit 1 and visit 3 (Wilcoxon signed‐rank test; p = 0.50).
Fig. 1.
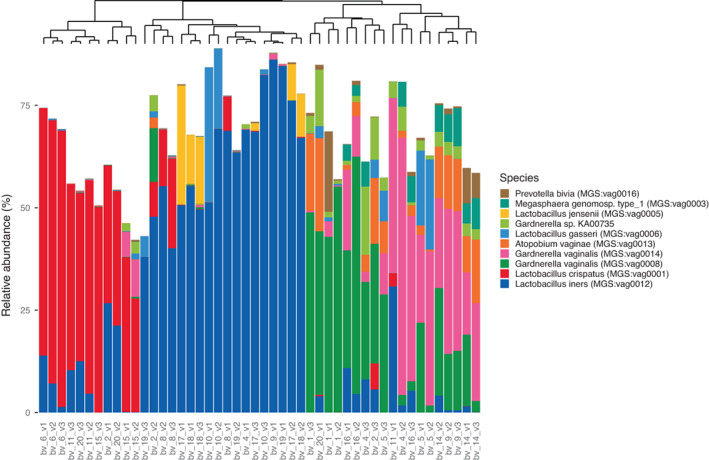
MGS abundance profiles of the top‐10 most dominant MGSs in the study vaginal samples. The samples are clustered based on Bray–Curtis dissimilarity and a phylogenetic tree on top of the plot exhibits the different species clusters. Bars do not sum to 100% because we represent only the 10 most dominant species across samples, and because some reads mapped to non‐MGS‐annotated genes.
Species detection and species abundance profiles in vaginal swabs
In the vaginal microbiota of the 16 women, we detected 25 individual species, of which six were Lactobacillus spp. Besides Lactobacillus spp., other dominant non‐lactobacillus species were especially Gardnerella vaginalis and Atopobium vaginae (all MGS are shown in Table S1). In Fig. 1, we show the annotated metagenomic species (MGSs) abundance profiles of the top‐10 most dominant vaginal MGSs. We observed three different sample clusters with one dominant species for each sample. The three predominant species G. vaginalis, L. iners, and L. crispatus characterize the three sample groups observed. Principal coordinates analysis (PCoA) (Fig. 2) based on Bray–Curtis dissimilarities among samples confirmed the existence of three different vaginal clusters and did not show any separation of samples between visits, indicating no effect of the probiotic treatment on the vaginal microbiome structure (confirmed by a Permanova test on all samples, testing the effect of visit stratification on the microbiome structure [R 2 = 0.02; p = 0.309]). Samples from the same subject generally showed limited compositional differences between timepoints, except for subjects 2, 4, 9, 11, and 20 who shifted from one group on visit 1 to another for sampling 2 and 3. Subjects 11 and 20 were initially dominated by G. vaginalis (at visit 1) and moved to a group dominated by L. crispatus (at visits 2 and 3). Yet, subjects 4 and 9 were dominated by L. iners (at visit 1) but moved to a group dominated by G. vaginalis (at visits 2 and 3). Other subjects with L. iners were all in the same group throughout the study. Subject 2 was the only participant to have three different groups based on PCoA (Fig. 2). To further confirm that the probiotics did not affect the vaginal microbiome, we tested for differentially abundant MGSs between baseline (visit 1) and during treatment (visit 2), and between baseline and post‐treatment (visit 3). There were no significant differentially abundant MGSs, either between visit 1 and visit 2 (FDR adjusted p > 0.88), or between visit 1 and visit 3 (FDR adjusted p > 0.23).
Fig. 2.
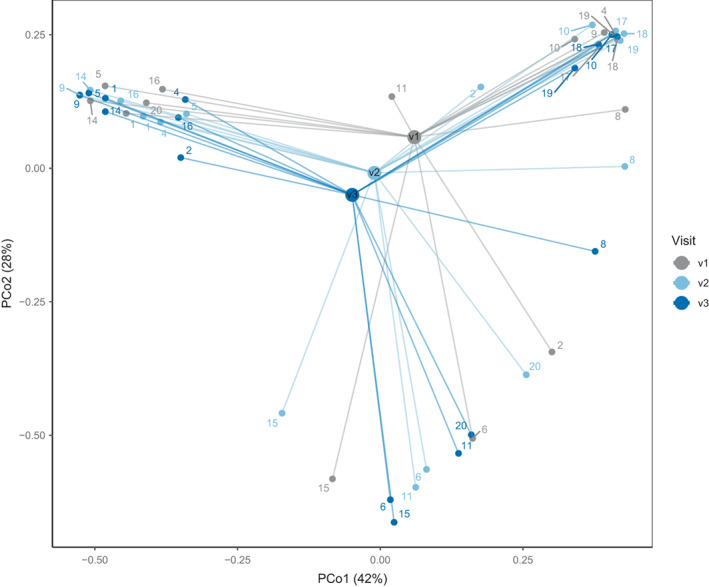
Vaginal samples. PCoA based on Bray–Curtis dissimilarities between vaginal samples calculated using the MGS abundances in vaginal samples. Numbers denote the subject, and the samples are color coded by visit. The median, of all the samples per visit, is expressed with a larger dot (centroid).
We also tested the differences in functional potential between the during‐ and post‐treatment time points (visits 2 and 3, respectively), and the baseline (visit 1) using summarized eggNOG groups and KEGG module abundances. We considered only orthologous groups and KEGG modules that were present in at least 10% of the samples, resulting in pruned tables of 7771 eggNOG groups and 466 KEGG modules. There were no differentially abundant eggNOG groups, either between visit 1 and visit 2 (all FDR‐adjusted p = 1) or between visit 2 and visit 3 (all FDR‐adjusted p > 0.91). There were also no differentially abundant KEGG modules, either between visit 1 and visit 2 (all FDR‐adjusted p = 1) or between visit 2 and visit 3 (all FDR‐adjusted p > 0.94). These results indicate that the oral probiotic had no significant effects on the microbial functional potential of the vaginal microbiota.
DISCUSSION
The purpose of this study was to investigate, if lactobacilli from oral probiotics had any impact on the vaginal microbiome and its functional potential in 16 healthy, non‐antibiotic treated Danish women. All participants followed an oral probiotic treatment, and their microbial changes were compared with their baseline sample (visit 1). This approach enabled us to characterize the vaginal microbiome of healthy women in reproductive age confirming species and species clusters previously characterized in this niche [2, 3, 6, 7, 8]. Although L. gasseri was detected in the vaginal microbiota of some study participants, this microbe was an endogenous L. gasseri strain and not the probiotic strain. Previous publications have shown vaginal colonization by L. rhamnosus following oral administration, which do not agree with our results. This could be due to a too low probiotic dose, unsuccessful translocation of the probiotic through the gastrointestinal route or signals below the detection levels.
We found three different vaginal clusters and did not show any separation of samples between visits, indicating no effect of the probiotic treatment on the vaginal microbiome structure. Overall, samples from the same subject generally showed limited compositional differences between timepoints, except for a few subjects who changed dominating species of their vaginal microbiome between sampling. Yet, the changes in these subjects were heterogenous with no indication of a specific influence from the probiotic strains. Furthermore, we found no indications that the oral probiotic had any significant effects on the microbial functional potential, since there were no differentially abundant eggNOG groups or differentially abundant KEGG modules, either between visit 1 and visit 2 or between visit 2 and visit 3. Finally, we observed no statistically significant taxonomical changes in the vaginal microbiome between visits.
Lactobacilli administered orally or intra‐vaginally have been tested for their effectiveness in preventing the recurrence of bacterial vaginosis (BV) [4, 5]. Oral administration has been introduced in a simple attempt to use lactobacilli via a more practical route as compared with the direct administration [12]. Randomized clinical trials have suggested that intra‐vaginal administration of lactobacilli helped to cure women with BV more frequently than administration of a placebo or no treatment [4, 5]. We have too few women in this study to confirm or reject these results. Yet, our results prove the need for appropriate randomized clinical trials in order to investigate the effect of oral probiotic on the structure of the vaginal microbiome and the functional potential in women suffering from vaginosis, with and without antibiotic treatment, as well as on healthy individuals.
Our study had several other limitations that may have influenced the outcome. Firstly, we had a limited number of participants, and the period was 2 months, within which only 3 vaginal swabs were taken. Secondly, all participants performed the vaginal swabbing with no control of how it was performed, nor did we control time from the last day of menses or have any detail of the cycle of the menstrual phase. However, no women took swabs during menstruation. Furthermore, we did not include a control group and did not have access to participants´ data to build a phenotypic profile. We chose to use swabs as sampling was performed by participants, as cytobrush and swab sampling has been found to provide a comparable microbiome composition and detect a small proportion of underlying species [13]. We have too few women in this study to confirm or reject results of previous studies, although we are convinced our method of detection is more meticulous. Several studies have illustrated that colonization by species such as L. crispatus, L. gasseri, L. iners, and L. jensenii is essential for a healthy vaginal microbiota, whereas L. rhamnosus and L. reuteri are commonly used as ingredients of oral probiotics but are species more prone to colonize the gut, and as transient colonizers of the vagina only [14]. We cannot rule out, that our results had been different had we used a strain of a Lactobacillus spp. known to colonize the vagina and not L. rhamnosus [2, 3, 5]. Of note, the strains used here had been isolated from human vaginal swap samples. Of note, in a randomized placebo‐controlled study including 100 women, Larsson et al. showed that the vaginal installation of Lactobacillus gasseri EB01‐DSM 14869 and Lactobacillus rhamnosus PB01‐DSM 14870 (EcovagFlora™), lengthened the time to BV relapse significantly, why intra‐vaginally administration may be beneficial, but is beyond the scope of our study [15]. Nonetheless, our conclusion agrees with Birte J. Wolff et al. that on young, self‐reported healthy women, the oral probiotic had no detectable effect on either the composition or the functional potential of the vaginal microbiota.
Future clinical studies would benefit from administering the probiotics during conditions of low bacterial presence, for example, during/after antibiotic treatment, or on other occasions where microbial vaginal imbalance would occur. We would also recommend a clinical trial with increased number of study participants, and the probiotic administered by the vaginal route to test whether these strains can thrive and colonize the vaginal niche.
CONCLUSION
Thus, the results indicate that the oral administration of the probiotics did not cause changes taxonomically or functionally in the vaginal microbiome. Although few subjects changed vaginal microbiome composition clusters, there were no significant taxonomical or functional changes associated to the treatment, suggesting an overall stable microbiota during the study timeframe. Thus, through shotgun sequencing, we could not reliably confirm that the orally administered probiotic strains have any indirect or direct impact on the vaginal microbiome of healthy women.
FUNDING
Exclusively funded by Deerland Probiotics and Enzymes A/S. Deerland Probiotics & Enzymes A/S was the primary financial contributor for the project and covered all expenses including, vaginal swabs, compensation for the participants, the microbiological analyses, and assessment of the generated data. The funding agencies had no role in designing and conducting the study, analysis and interpretation the data, or the writing, review, and approval of the manuscript.
CONFLICTS OF INTEREST
FBH and NFM are medical doctors employed by the public health care system in Denmark. FBH and NFM declare no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous three years, no other relationships or activities that could appear to have influenced the submitted work. LSM and EB are employed by Deerland Probiotics and Enzymes A/S a commercial company producing and selling probiotics, including probiotics containing Lactobacillus spp. Deerland Probiotics & Enzymes A/S had no role in conducting the study, analysis or interpretation the data. JBH and AP are employed by Clinical Microbiomics, a private company. Clinical Microbiomics have been involved in the study to ensure correct microbiological analyses. Clinical Microbiomics have had no influence on the purpose.
TRANSPARENCY DECLARATION
All authors affirm that this manuscript is conducted honest, accurate, and transparent.
Supporting information
Table S1 All species detected in the vaginal samples.
Table S2 Strain‐level analysis of Lactobacillus gasseri in during‐treatment and post‐treatment samples.
Table S3 Primer sequences and amplified DNA fragments.
Table S4 Concentration of selected primers and probes used in individual reaction mixtures.
Table S5 qPCR cycling program for each qPCR reaction mixture. The qPCR amplification was performed on a MyGo Pro instrument (IT‐IS Life Science Ltd.).
Table S6 Volume of primers, probes, DNA templates for amplification and qPCR reagents (Ampliqueen) in each qPCR reaction mixture.
References
- 1. Keith JW, Pamer EG. Enlisting commensal microbes to resist antibiotic‐ resistant pathogens. J Exp Med. 2019;216(1):10–19. 10.1084/jem.20180399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith SB, Ravel J. The vaginal microbiota, host defence and reproductive physiology. J Physiol. 2017;595(2):451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falagas ME, Betsi GI, Athanasiou S. Probiotics for the treatment of women with bacterial vaginosis. Clin Microbiol Infect. 2007;13(7):657–64. [DOI] [PubMed] [Google Scholar]
- 5. Larsson P‐G, Brandsborg E, Forsum U, Pendharkar S, Andersen K, Nasic S, et al. Extended antimicrobial treatment of bacterial vaginosis combined with human lactobacilli to find the best treatment and minimize the risk of relapses. BMC Infect Dis. 2011;11(1):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Younes JA, Lievens E, Hummelen R, van der Westen R, Reid G, Petrova MI. Women and their microbes: the unexpected friendship. Trends Microbiol. 2018;26(1):16–32. [DOI] [PubMed] [Google Scholar]
- 7. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive‐age women. Proc Natl Acad Sci USA. 2011;108(Suppl):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Albert AYK, Chaban B, Wagner EC, Schellenberg JJ, Links MG, Van SJ, et al. A study of the vaginal microbiome in healthy Canadian women utilizing cpn 60‐based molecular profiling reveals distinct Gardnerella subgroup community state types. PLoS One. 2015;10(8):e0135620. 10.1371/journal.pone.0135620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reid G, Charbonneau D, Erb J, Kochanowski B, Beuerman D, Poehner R, et al. Oral use of lactobacillus rhamnosus GR‐1 and L. fermentum RC‐14 significantly alters vaginal flora: randomized, placebo‐controlled trial in 64 healthy women. FEMS Immunol Med Microbiol. 2003;35(2):131–4. [DOI] [PubMed] [Google Scholar]
- 10. Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32(8):822–8. [DOI] [PubMed] [Google Scholar]
- 11. Munk P, Andersen VD, de Knegt L, Jensen MS, Knudsen BE, Lukjancenko O, et al. A sampling and metagenomic sequencing‐based methodology for monitoring antimicrobial resistance in swine herds. J Antimicrob Chemother. 2017;72(2):385–92. [DOI] [PubMed] [Google Scholar]
- 12. Homayouni A, Bastani P, Ziyadi S, Mohammad‐Alizadeh‐Charandabi S, Ghalibaf M, Mortazavian AM, et al. Effects of probiotics on the recurrence of bacterial vaginosis: a review. J Low Genit Tract Dis. 2014;18(1):79–86. [DOI] [PubMed] [Google Scholar]
- 13. Mitra A, MacIntyre DA, Mahajan V, Lee YS, Smith A, Marchesi JR, et al. Comparison of vaginal microbiota sampling techniques: Cytobrush versus swab. Sci Rep. 2017;7(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chee WJY, Chew SY, Than LTL. Vaginal microbiota and the potential of lactobacillus derivatives in maintaining vaginal health. Microb Cell Fact. 2020;19(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsson P‐G, Stray‐Pedersen B, Ryttig KR, Larsen S. Human lactobacilli as supplementation of clindamycin to patients with bacterial vaginosis reduce the recurrence rate; a 6‐month, double‐blind, randomized, placebo‐controlled study. BMC Womens Health. 2008;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 All species detected in the vaginal samples.
Table S2 Strain‐level analysis of Lactobacillus gasseri in during‐treatment and post‐treatment samples.
Table S3 Primer sequences and amplified DNA fragments.
Table S4 Concentration of selected primers and probes used in individual reaction mixtures.
Table S5 qPCR cycling program for each qPCR reaction mixture. The qPCR amplification was performed on a MyGo Pro instrument (IT‐IS Life Science Ltd.).
Table S6 Volume of primers, probes, DNA templates for amplification and qPCR reagents (Ampliqueen) in each qPCR reaction mixture.


