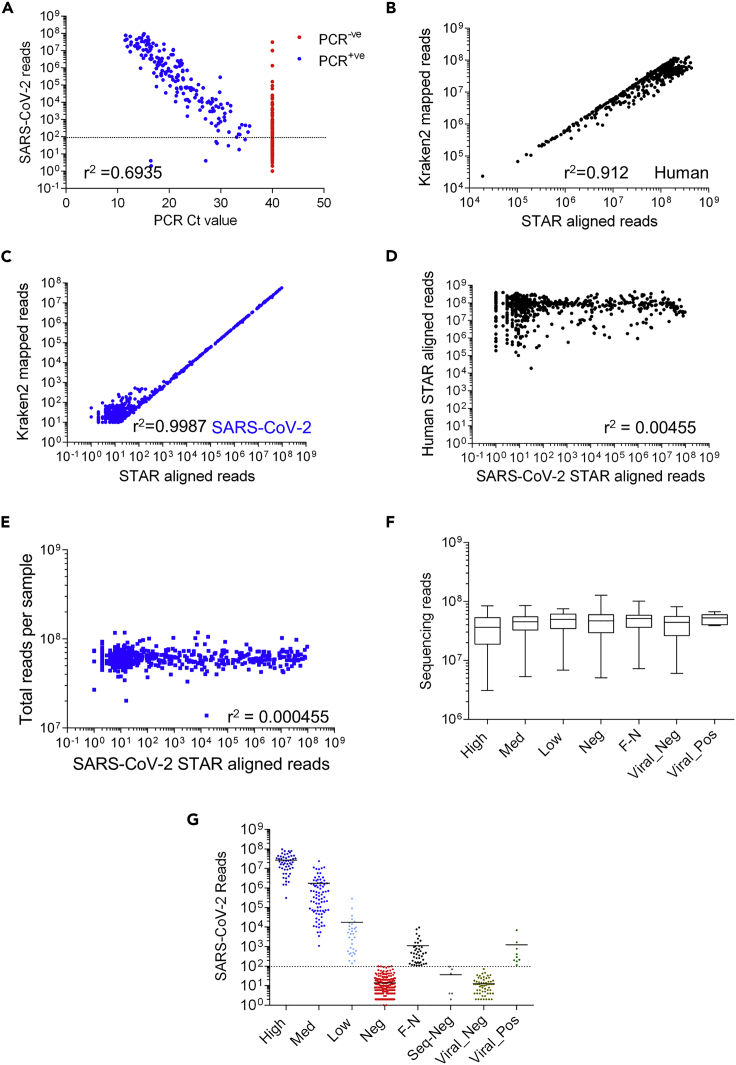
Figure 1.
Comparison of Human and SARS-Cov-2 aligned reads from nasal pharyngeal swabs
(A) Comparison of PCR Spike gene Ct values for samples vs STAR SARS-CoV-2 aligned reads. A PCR Ct value of 40 was defined as PCR negative (PCR -ve: red, 474 samples) and a Ct value below 40 was defined as positive (PCR + ve: blue, 194 samples). The dotted line denotes a sequencing cut off value of 100 reads. Pearson correlation of PCR-positive samples (Ct values vs log10 number of Sequencing reads) yields a slope of −3.7 and r2 value of 0.6935.
(B) Comparison of Kraken2 and STAR aligned reads to the human reference genome (GRCh38). Pearson correlation of log10 transformed kraken vs STAR aligned reads to GRCh38 yields a slope of 0.89 and r2 value of 0.912.
(C) Comparison of Kraken2 and STAR aligned reads to the SARS-CoV-2 reference genome (Wuhan strain). Pearson correlation of log10 transformed kraken vs STAR aligned reads to SARS-CoV-2 genome yields a slope of 0.88 and r2 value of 0.978.
(D) Comparison of STAR aligned reads to the SARS-CoV-2 vs. human reference Genome (GRCh38) each point represents one sample. Pearson correlation of log10 transformed data yields a slope of 0.019 and r2 value of 0.00455.
(E) Comparison of STAR aligned reads to the SARS-CoV-2 reference genome vs. the total number of reads in the sample (sequencing depth). Pearson correlation of log10 transformed data yields a slope of 0.0008 and r2 value of 0.000455 (F). Total number of reads obtained from all samples split by SARS-CoV-2 status based on PCR and Sequencing results. There were no significant differences between sample read number across categories (1-way ANOVA with post hoc Bonferroni’s test). Data shown are median (min to max values) of 52, 81, 33, 275, 42, 66 & 9 samples, respectively.
(G) Separation of samples into categories by PCR (High: Ct ≤ 18, Med: 18 < Ct ≤ 24, Low: 24 < Ct < 40, Neg: Ct ≥ 40) and Kraken2 (Viral) values. We used a total aligned reads values of 100 as our cutoff for positive / negative identification of SARS-CoV-2 by sequencing (dotted line). Samples that were PCR negative and sequencing positive were designated as false negative (F–N) samples. Group names indicate SARS-CoV-2 PCR status. (Of note 40 samples had zero values and are not shown). Samples with other viral infections, based on Kraken2 identification of other respiratory virus infection were removed from subsequent analysis. Data shown are individual values of 52, 81, 33, 275, 42, 66, & 9 samples, respectively (see Figure S1A).