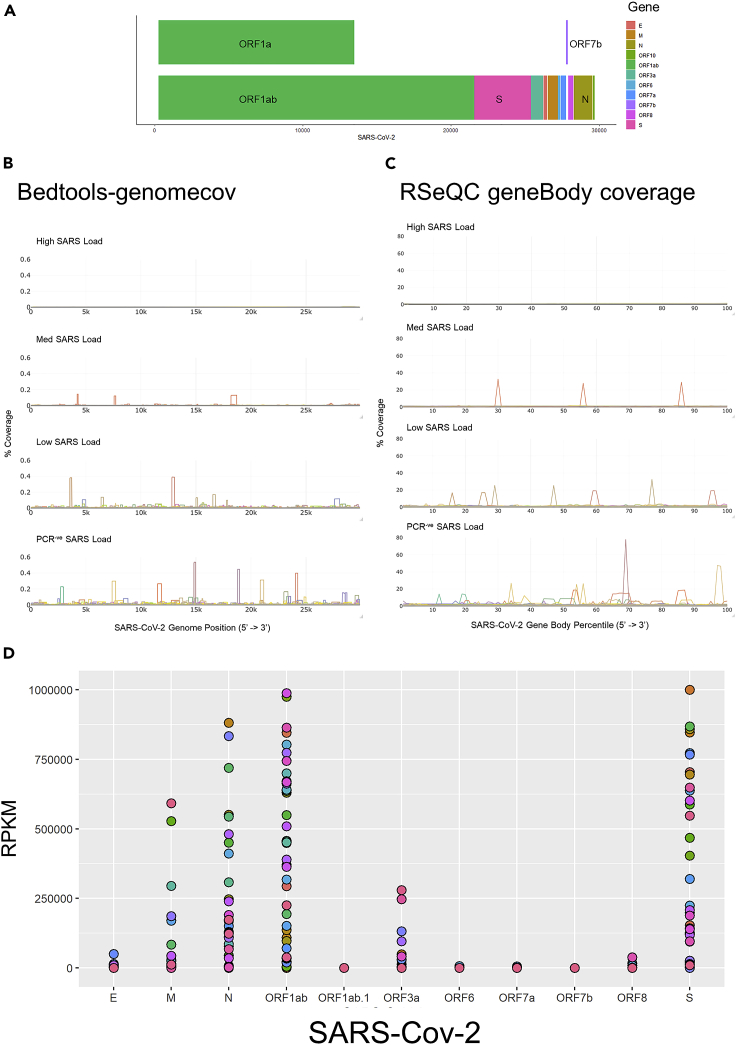
Figure 2.
Analysis of alignment of data to SARS-CoV-2 genome
(A) Schematic overview of the SARS-CoV-2 genome.
(B) Comparison of SARS-CoV-2 genome coverage as determined using Bedtools genomecov. Data were partitioned based on viral load of PCR-positive samples (High, Med, Low), and the PCR negative sequencing positive samples (PCR-ve). % Coverage was calculated for each sample as ((number reads in SARS-CoV-2 genome position) / (total number of SARS-CoV-2 aligned reads))∗100.
(C) Comparison of SARS-CoV-2 transcript coverage as determined using RSeQC geneBody coverage. Data were partitioned based on viral load of PCR-positive samples (High, Med, Low), and the PCR negative sequencing positive samples (PCR-ve). % Coverage was calculated for each sample as ((number reads in SARS-CoV-2 gene body percentile) / (total number of SARS-CoV-2 aligned reads))∗100.
(D) Expression of SARS-CoV-2 genes in RNA sequencing data. Read counts were converted to TPM and plotted for each PCR negative sequencing positive sample.