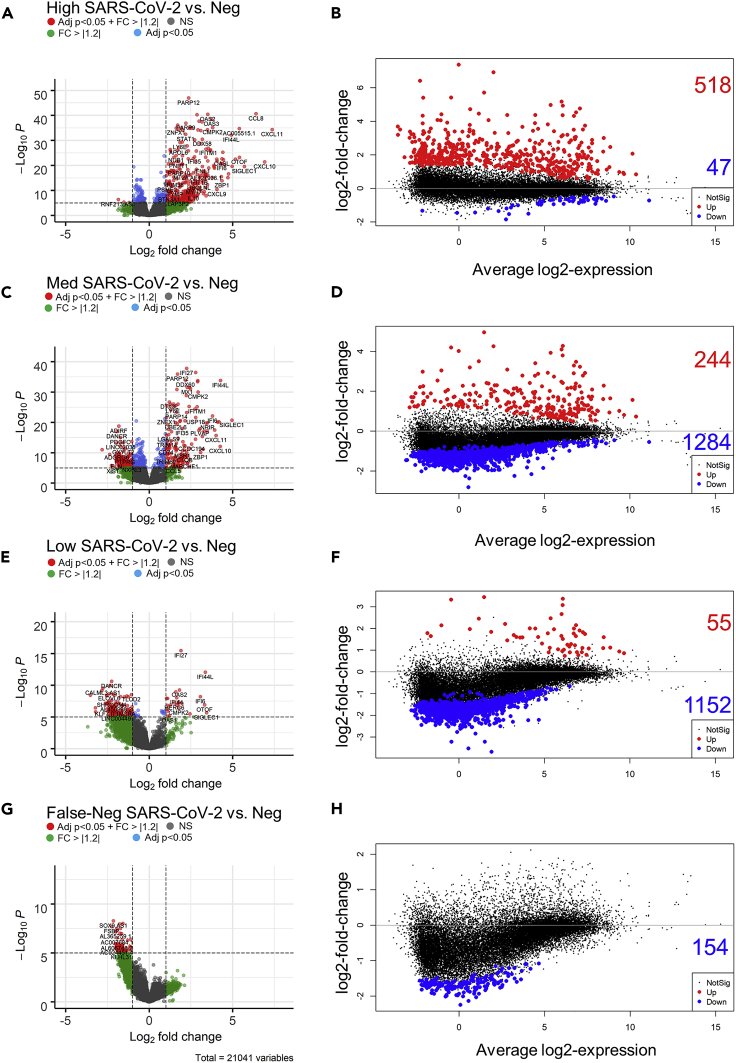
Figure 3.
Gene expression in nasal pharyngeal swabs from patients exposed to SARS-CoV-2 based on high, medium, low and (F–N) viral load vs. uninfected patients (PCR and sequence negative samples, Neg)
(A, C, E and G) Expression depicted in volcano plots. Volcano plots show the raw unadjusted p value, and the values that pass our inclusion criteria are depicted in red (Differentially expressed genes (log2 fold change > 1.2 or < −1.2 and adjusted p < 0.05 (FDR)). The dotted lines show the cut off raw p (adjusted p value = 0.05 unadjusted p of approximately 1 × 10−5) and log2-fold change (±1.2) values.
(B, D, F and H) MA plots showing average log2 expression vs. log2-fold change of up (red) and down (blue) regulated genes. Non-significant changes in expression are shown in black. Note the shift from predominantly increased gene expression in high viral load patients, to decreased gene expression in low viral load patients.