FIGURE 2.
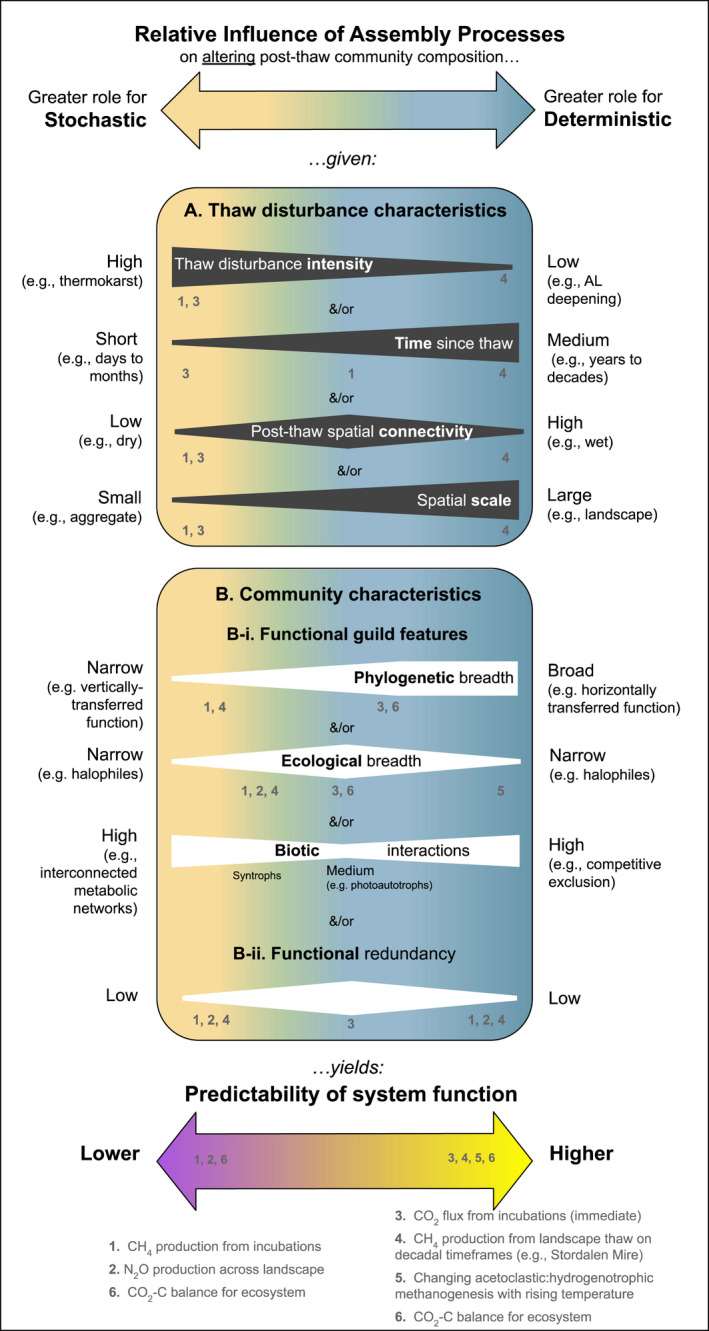
Conceptual framework connecting thaw disturbance and community characteristics to assembly and the predictability of system function. The relative contribution of stochastic and deterministic assembly processes are expected to be impacted by permafrost thaw disturbance characteristics (e.g., time since thaw and thaw disturbance intensity) and functional guild features (phylogenetic and ecological breadth) discussed in Sections 2, 3, 4. Note that all grey and white wedges are independent and are visual representations of the scale of each disturbance or community characteristic (e.g., when disturbance intensity is high the wedge is big, and when it is low the wedge is small). Phylogenetic breadth refers to functional guild evolutionary cohesiveness (e.g., narrow for anaerobic methane oxidation, broad for aerobic methane oxidation); phylogenetically narrow, for example, indicates that functional guild members (e.g., those members sharing a trait) are phylogenetically clustered (see Section 4.1). Ecological breadth among members of a functional guild refers to how similarly members respond to environmental drivers (i.e., an ecologically narrow guild responds more similarly to a change in abiotic and/or biotic conditions). Notably, these operational concepts depend on the trait and habitat characteristic being considered (e.g., the guild of anaerobic methane oxidizers is phylogenetically narrow and ecologically narrow with respect to oxygen, but not to salinity). See Section 2. The numbers indicate observations in the field or laboratory (references follow), and their placement on the wedges shows the relative contribution of disturbance and functional guild features that affect whether stochasticity or determinism dominate. These characteristics have implications for the predictability of community function (bottom arrow) arising from dominance by stochastic versus deterministic processes, and thus the approaches required to model specific microbiome outputs (such as greenhouse gas emissions). References: 1. McCalley et al., 2014; 2. Drake et al., 2015; Spencer et al., 2015; 3. Allan et al., 2014; 4. See example below; 5. Knoblauch et al., 2018, Ernakovich et al., 2017; 6. Siljanen et al., 2019.
