FIGURE 1.
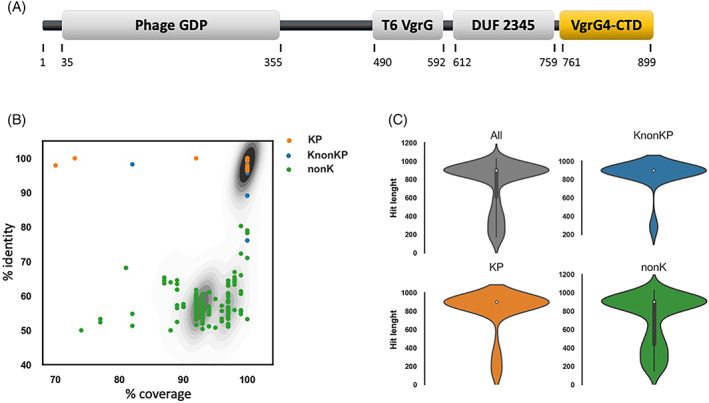
VgrG4‐CTD sequence does not contain known domains but is conserved in several K. pneumoniae isolates (KP), Klebsiella non‐K. pneumoniae (KnonKP), and also non‐Klebsiella bacteria (nonK). (A) Schematic representation of the VgrG4 amino acid sequence. Functional domains according to NCBI Conserved Domain Database are represented as gray boxes: Phage GPD, T6SS VgrG, and DUF 2345. The C‐terminal region (amino acids 761–899) does not contain any known domain, and is highlighted in a yellow box (VgrG4‐CTD). (B) Scatter plot showing the % identity, and % coverage of Blastp hits of VgrG4‐CTD. Contour plot in the background represents the density of hits falling in the same position of the plot. (C) Violin plots showing the density of VgrG4‐CTD hits according to hit length (amino acids).
