Figure 7.
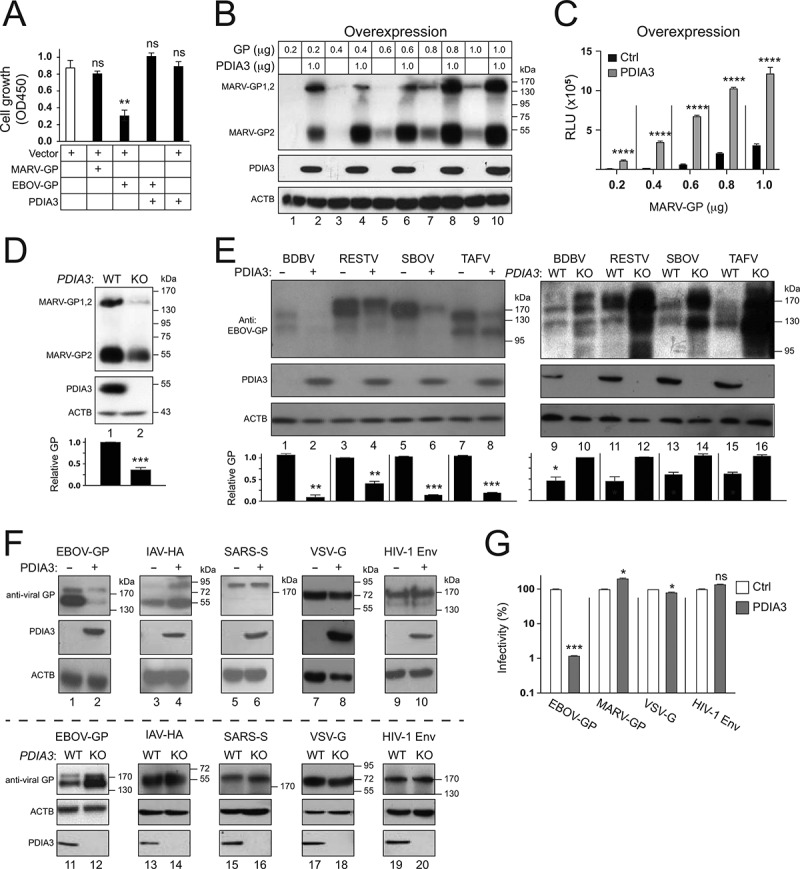
Specificity of the PDIA3 inhibitory activity on viral glycoprotein expression. (a) HEK293T cells were transfected with a control (Ctrl) vector, or vectors expressing MARV-GP, EBOV-GP, and/or PDIA3. After 48 h, cells were incubated with WST-8 and viable cells were counted by a microplate reader with a 450 nm filter. Results are presented as optical density (OD450). (b) HEK293T cells were transfected with increasing amounts of MARV-GP with a HiBiT-tag and 1.0 μg PDIA3 expression vectors. The GP expression was determined by Nano-Glo® HiBiT Blotting System. (c) MARV-GP expression in B was further quantified by Nano-Glo® HiBiT Lytic Reagent and are presented relative luminescence units (RLU). (d) MARV-GP was expressed in HEK293T WT and PDIA3-KO cells, and its expression was determined by the HiBiT Blotting system. (e) GPs from indicated ebolaviruses were expressed with PDIA3 in HEK293T cells. Alternatively, they were expressed in HEK293T WT and PDIA3-KO cells. GP expression was determined by WB using anti-EBOV-GP. (f) Indicated viral glycoproteins were expressed with PDIA3 in HEK293T cells. Alternatively, they were expressed in HEK293T WT and PDIA3-KO cells. Their expression was analyzed by WB using their specific antibodies. (g) HIV-1 firefly luciferase reporter viruses with authentic HIV-1 Env or pseudotyped with EBOV-GP, MARV-GP, or VSV-G were produced from HEK293T cells in the presence of ectopic PDIA3. The entry of viruses with HIV-1 Env was determined in TZM-bI cells, and that of pseudoviruses was determined in HEK293T cells. Viral entry is shown as relative values, with the value in the absence of PDIA3 normalized to 100%. The levels of GP in D and E were further quantified. Error bars in A, C, D, and G represent SEMs calculated from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns (p > 0.05).
