Figure 2.
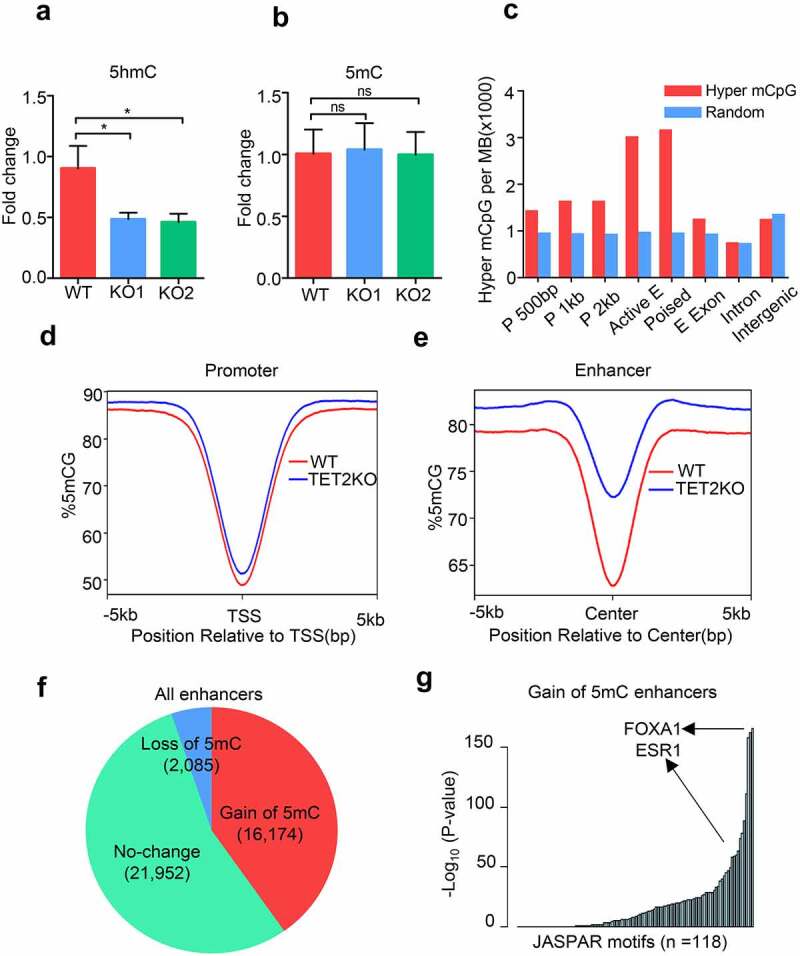
TET2 depletion results in aberrant DNA methylation within a subgroup of enhancers. (a-b) HPLC/MS analysis of the global 5hmC (a) and 5mC (b) levels in WT and TET2 KO cells. (c) Histogram plots showing the relative enrichment of the hypermethylated CpG sites at several genomic features. ‘P 500bp’ indicates promoter defined as transcription start site ± 500 bp. ‘Active and Poised E’ denotes active and poised enhancer were defined with public H3K4me1 and H3K27ac ChIP-seq data. Random consists of random samplings of genomic loci. (d-e) Profile plot of the average 5mC signal on all promoters (d) and all enhancers (e) in WT (red line) and TET2 KO (blue line) MCF7 cells. (f) Pie chart showing the percentage of ‘gain of 5mC,’ ‘loss of 5mC’ and ‘no change’ enhancers in TET2 KO MCF7 cells. (g) Representation of TF binding motifs overrepresented on the ‘gain of 5mC’ enhancers. Human promoters were used as the comparison library.
