Figure 1.
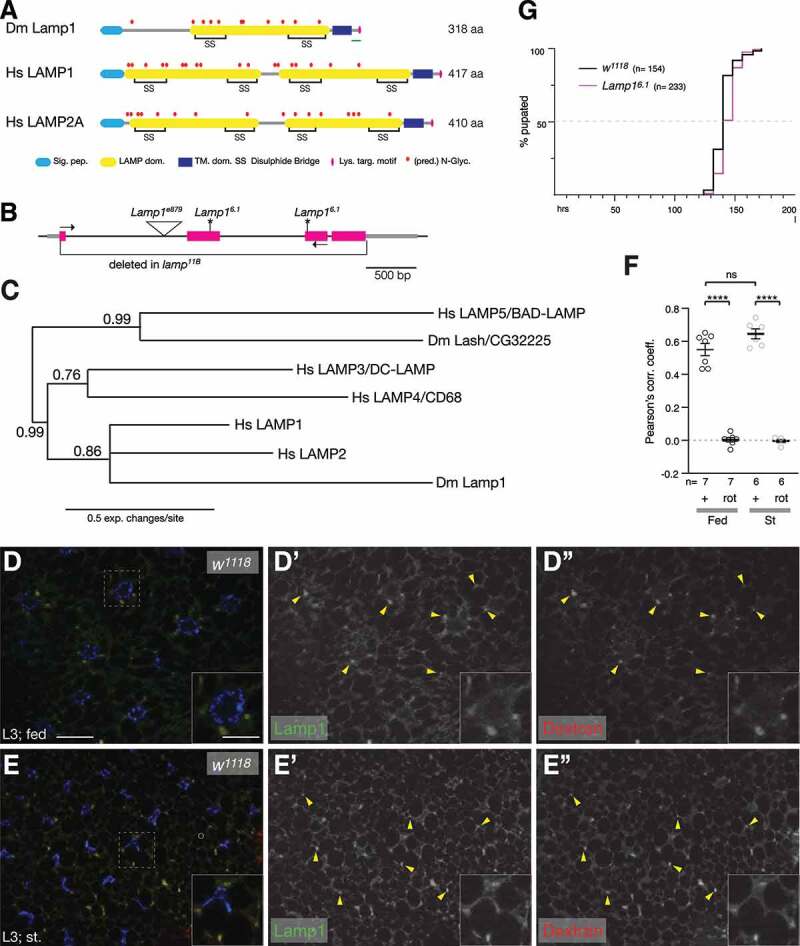
Lamp1 mutants are viable and have no developmental delay. (A) Schematic comparing Drosophila Lamp1 with human LAMP1 and LAMP2A. Note that Lamp1 has only one lumenal LAMP domain. The green bar indicates the peptide region used to raise the Lamp1 antibody. (B) Schematic of Lamp1 locus with mutant alleles indicated. UTRs are in gray, coding sequence in magenta. Lamp16.1 contains two frame shifts (*) at the positions of both gRNAs used to induce it. Arrows depict location of RT-PCR primers. (C) Bayesian phylogenetic tree of indicated LAMP proteins. Numbers indicate bootstrap values. (D, E) In 3rd instar larval fat body under fed (C) and starved conditions (D), Lamp1 colocalizes with TRITC-Dextran-labeled endolysosomes (examples marked by yellow arrowheads). Single channel images show Lamp1 (D’, E’) and Dextran (D”, E”), respectively. Insets show enlarged section indicated by dotted squares. Nuclei are in blue. Scale bars: 20 µm (10 µm in insets). (F) Pearson’s correlation coefficient for the colocalization of Lamp1 with TRITC-Dextran under indicated conditions. As control, one channel was rotated (rot) by 180°. One-way ANOVAs (Tukey correction) P < 0.0001; ****, P < 0.0001; ns, not significant. (G) Quantification of pupation timing shows that Lamp16.1 mutants have no developmental delay. n indicates total number of pupae scored.
