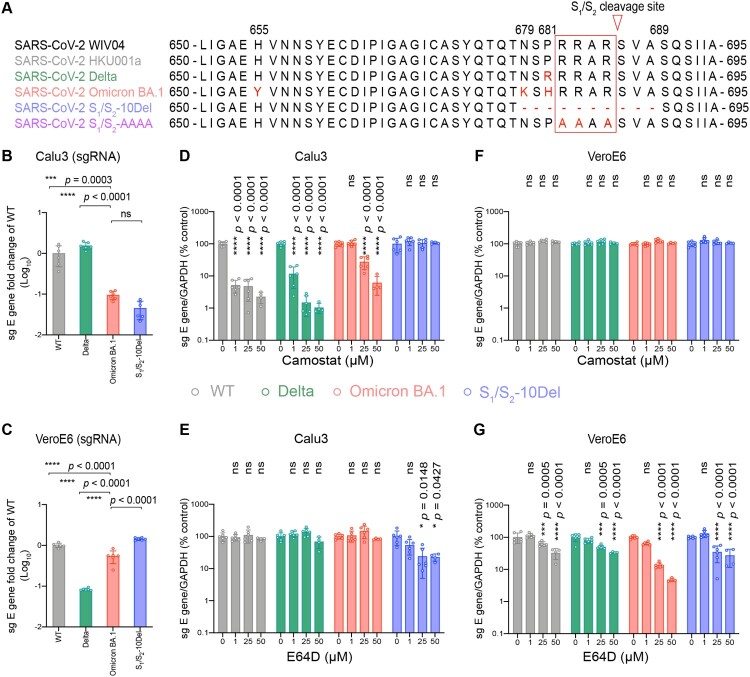
Figure 1.
Omicron BA.1 is less dependent on TMPRSS2-mediated plasma membrane entry pathway and is highly dependent on the endosomal entry pathway for virus entry. (A) Amino-acid sequence alignment of residues around the S1/S2 cleavage site of SARS-CoV-2 reference strain WIV04, HKU001a, Delta, Omicron BA.1, S1/S2-10Del, and S1/S2-AAAA. Amino acid positions are designated based on SARS-CoV-2 reference strain. (B–C) Calu3 and VeroE6 cells were challenged with SARS-CoV-2 WT, Delta, Omicron BA.1, or S1/S2-10Del. Cell lysates were harvested at 24 hpi for quantification of the subgenomic RNA of the envelope (sgE) gene (n = 6). (D–G) Calu3 and VeroE6 cells were pre-treated with indicated concentrations of camostat or E64D for 2 h followed by the authentic SARS-CoV-2 variants infection. The amount of viral subgenomic envelope RNA in harvested cell lysates at 24 hpi was determined by qRT-PCR (n = 4 for 50uM of camostat and E64D groups, n = 6 for the other groups). Error bars were calculated by using log-transformed data and represented mean ± SD from the indicated number of biological repeats. Statistical significances were determined with one way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant; WT, wildtype SARS-CoV-2.