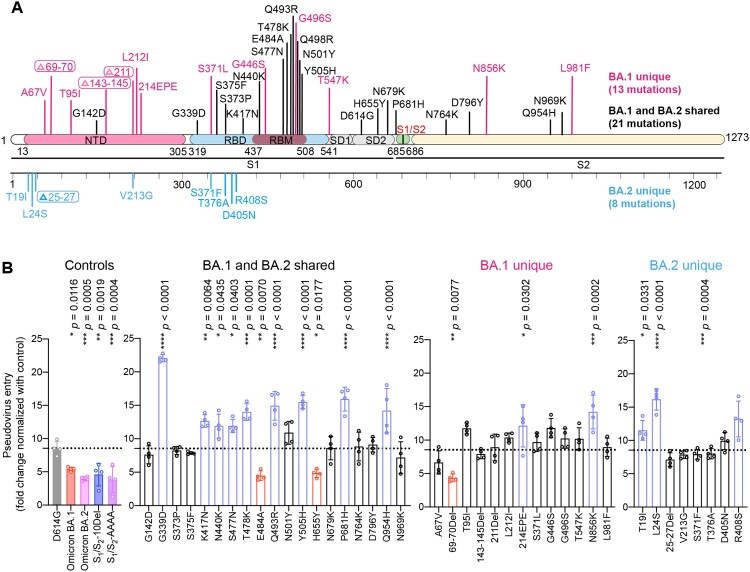
Figure 2.
Spike determinants for the reduced TMPRSS2 usage of Omicron BA.1 and BA.2. (A) Schematic of all amino acid mutation sites on Omicron BA.1 and BA.2 spike when compared to ancestral SARS-CoV-2 spike. (B) 293T cells were transfected with hACE2 or co-transfected with hACE2 and TMPRSS2, followed by transduction with pseudoviruses bearing the spike of SARS-CoV-2 D614G, Omicron BA.1, Omicron BA.2, S1/S2-10Del, S1/S2-AAAA and individual mutation at 24 h post-transfection. Pseudovirus entry was quantified by measuring the luciferase signal (n = 4). Fold changes in the luciferase signal were normalized to the mean luciferase readouts of cells with only hACE2 overexpression. Data represent mean ± SD from the indicated number of biological repeats. Statistical significance was determined with one way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant.